
Personalized antihypertensive treatment guided by pharmacogenomics in China
Introduction
Hypertension is a leading cause of disability and cardiovascular mortality. According to the China Cardiovascular Disease Report 2018, hypertension is highly prevalent among Chinese adults, occurring at a rate of 27.9% in the population of 245 million patients, with only 16.8% of patients achieving blood pressure (BP) control (1). Antihypertensive drugs have long been adopted as the main approaches for reducing BP to physiologically normal levels and preventing hypertension-associated target organ damage. Despite the availability of effective antihypertensive drugs, the rate of BP control remains poor due to multiple factors, including irrational prescription of antihypertensive drugs due to insufficient clinical experience and apparent drug resistance (2). Major evidence-based clinical guidelines fundamentally use a standard universal approach to prescribe antihypertensive drugs which, at the individual patient level, are not highly personalized. Clinical practices have shown the heterogeneity of patients’ responses to antihypertensive drugs. Genetic polymorphisms and pharmacokinetic characteristics may partially account for the interindividual variability (3,4). Increasing evidence supports the establishment of personalized pharmacotherapeutic treatment based on a patient’s genetic background. Personalized pharmacotherapy based on genetic information is therefore expected to become the principal mode of future antihypertensive treatment especially in non-responsive hypertensive patients, in order to distinguish which kind of antihypertensive drug will lower BP most effectively, including economically, for individual patients. To date, the genetic research of hypertension has mainly focused on the causal genes of hypertension and the mechanisms of BP elevation, such as the angiotensin converting enzyme insertion/deletion (ACE I/D) (5) polymorphisms and ATPase plasma membrane Ca2+ transporting 1 (ATP2B1) (6) polymorphisms which were discovered to be associated with high BP. However, there are no currently available clinical tests to guide the choice of antihypertensive drugs, and the association between gene polymorphisms and increased responsiveness to antihypertensive drugs, including angiotensin I-converting enzyme inhibitors (ACEIs), angiotensin II receptor blockers (ARBs), beta blockers, calcium channel blockers (CCBs) and diuretics remains controversial (7). Thus, our study analyzed the distribution of antihypertensive drug-related gene polymorphisms, in the Changsha county of Hunan province. A polymerase chain reaction (PCR)-fluorescence probe was used to detect the 7 genetic polymorphic loci (CYP2D6*10, ADRB1, CYP2C9*3, AGTR1, ACE, CYP3A5*3 and NPPA) associated with the effect of antihypertensive drugs, and then the antihypertensive drug doses and type were adjusted based on the gene phenotype and pharmacokinetic characteristics of different patients or based on the clinical experience of the designated doctor. By comparing the difference of curative effect between these 2 groups, we aimed to investigate the value of genotype-based prescribing in clinical antihypertensive therapy, and to provide a potential approach to the personalized management of hypertension.
Methods
Participants
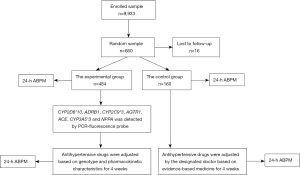
A total amount of 9,933 hypertensive patients aged from 26 to 96 years registered in township hospitals and street health service centers from 2017 to 2018 in the Changsha County of Hunan province were enrolled by the study’s investigators, with the patients’ written informed consent. The participants comprised 4,015 males and 5,918 females of average age 64.16±8.76 years. The eligibility criteria comprised all of the following: (I) people aged >18 years old, (II) a diagnosis of chronic hypertension (treated or untreated), and (III) according to the Revised Guidelines for the Prevention and Treatment of Hypertension in China 2010, having a systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg three times on different days (8). The exclusive criteria included any 1 of the following: (I) secondary hypertension, (II) acute cardiovascular and cerebrovascular events within the previous 3 months, and (III) severe cardiomyopathy, rheumatic heart disease, congenital heart disease, or severe liver and kidney dysfunction. Then, 660 patients with hypertension were recruited from 20 township hospitals and street health service centers using a multistage cluster sampling design and were randomly divided into 2 groups at a ratio of 3:1. The experimental group received antihypertensive drug therapy adjusted according to the gene detection, and the control group received antihypertensive drug therapy according to evidence-based medicine. In the end, 16 patients were lost to follow-up, leaving 484 cases in the experimental group and 160 cases in the control group.
Clinical data collection
Gene samples were collected from the oral mucosa of the 9,933 hypertensive patients, and PCR-fluorescence probe technology was used to detect the 7 genetic polymorphic loci (CYP2D6*10, ADRB1, CYP2C9*3, AGTR1, ACE, CYP3A5*3 and NPPA) related to the metabolism, transport and target of 5 main kinds of antihypertensive drugs, including ACEIs, ARBs, beta blockers, CCBs and diuretics, which determined the patient’s response to antihypertensives. According to the gene phenotype and pharmacokinetic characteristics of different individuals, the most sensitive antihypertensive drugs with the least adverse effects were prescribed in a single-dose regime or combination therapy to the experimental group for 4 weeks, and the dosage adjustment of antihypertensives was based on the following principles: doubling the standard dose when the hypertension was moderately sensitive to certain antihypertensive drugs, and using the minimum dose to start treatment when the hypertension was highly sensitive to certain antihypertensive drugs (as shown in Table S1). In the control group, treatment was prescribed by the designated doctor based on the Revised Guidelines for the Prevention and Treatment of Hypertension in China 2010 (as shown in Table S2) for 4 weeks. Several adverse events were reported during the whole process, including mild ankle edema in 5 patients and fatigue in 6 patients. However, the side effects of the antihypertensive drugs disappeared after adjustment of medications and did not cause any cases to withdraw from the study. We used 24-hour ambulatory blood pressure monitoring (ABPM) to assess the BP level of both groups, pre- and post-intervention. The BP treatment target was 24-hour mean BP <130/80 mmHg, daytime mean BP <135/85 mmHg and night mean BP <120/70 mmHg, in accordance with recommendation from the American Heart Association guidelines (9) (Figure 1).
24-hour ABPM
Validated 24-hour ABPM (Oscar2; SunTech Medical, Inc., Morrisville, NC, USA) was applied to measure ambulatory BP through a routine day. All individuals were taught to accurately record the time when waking, going to bed, taking medicine and engaging in other activities. Measurements were taken every 30 minutes during the daytime and every 60 minutes during the night-time. Valid recordings were of at least 20 hours duration and a minimum of 70% of the expected 24-hour readings were required to be valid. Means of 24-hour, daytime and night-time ambulatory SBP and DBP were calculated (10).
Statistical analysis
The frequency of each locus was calculated by the frequency counting method and confirmed by the Hardy-Weinberg equilibrium fit test. Quantitative data were expressed as the mean ± standard error (x ± s), differences between groups were compared using the independent-sample t-test, and pre- and post-treatment intra-group differences were compared using the paired t-test. Enumeration data were expressed as percentage (%), and the comparison between pre- and post-treatment intra-group was performed by the paired χ2 test, comparison between groups was performed by the χ2 test. A value of P<0.05 was considered statistically significant. According to the sample size and BP control rate of the 2 groups, the power calculation that determined the study design was 1, which meant that if there was any difference in the rate of BP control between the 2 group, we were 100% confident of finding it. The software SPSS 22.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analysis.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of Xiangya Hospital of Central South University (No. 20170124028) and informed consent was taken from all individual participants.
Results
The frequency distribution of each gene locus
Among the 7 genetic polymorphic loci related to the efficacy of 5 main kinds of antihypertensive drugs, CYP2D6*10 and ADRB1(1165G>C) were related to the beta blocker, CYP2C9*3 and AGTR1(1166A>C) were related to the ARBs, ACE(I/D) was related to the ACEIs, and CYP3A5*3 and NPPA(2238T>C) were related to the CCBs and diuretics, respectively. The distribution of these 7 genetic polymorphic loci from 9,933 cases in Changsha County is shown in Table 1. There were no significant differences between genders in the mutation rates of the 7 gene loci (Table 2; P>0.05). Similarly, there was no significant correlation between genotype and body mass index (BMI), SBP, DBP, and family history of hypertension (Table 3; P>0.05). There was a failure to detect certain loci in several samples, probably due to other types of gene mutation carried in these samples.
Table 1
| WW | WM | MM | W (%) | M (%) | |
|---|---|---|---|---|---|
| CYP2D6*10 | 2,127 | 4,259 | 3,532 | 42.92 | 57.08 |
| ADRB1(1165G>C) | 640 | 3,765 | 5,528 | 25.4 | 74.6 |
| CYP2C9*3 | 9,083 | 823 | 27 | 95.59 | 4.41 |
| AGTR1(1166A>C) | 8,891 | 977 | 63 | 94.45 | 5.55 |
| ACE(I/D) | 4,763 | 4,193 | 976 | 69.06 | 30.94 |
| CYP3A5*3 | 962 | 4,228 | 4,742 | 30.97 | 69.03 |
| NPPA(2238T>C) | 9,739 | 149 | 44 | 98.81 | 1.19 |
WW, homozygous wildtype; WM, heterozygous mutant; MM, homozygous mutant; W, wild type; M, mutant.
Table 2
| Gene loci | Male (n=4,015) | Female (n=5,918) | χ2 | P | |||
|---|---|---|---|---|---|---|---|
| W (%) | M (%) | W (%) | M (%) | ||||
| CYP2C9*3 | 95.88 | 4.12 | 95.39 | 4.61 | 0.029 | 0.865 | |
| ADRB1(1165G>C) | 24.62 | 75.38 | 25.92 | 74.08 | 0.045 | 0.832 | |
| AGTR1(1166A>C) | 94.60 | 5.40 | 94.35 | 5.65 | 0.006 | 0.938 | |
| CYP2D6*10 | 42.11 | 57.89 | 43.47 | 56.53 | 0.038 | 0.746 | |
| ACE(I/D) | 69.03 | 30.97 | 69.09 | 30.91 | 0.001 | 0.993 | |
| NPPA(2238T>C) | 98.89 | 1.11 | 98.75 | 1.25 | 0.008 | 0.927 | |
| CYP3A5*3 | 31.16 | 68.84 | 30.84 | 69.16 | 0.002 | 0.961 | |
W, wild type; M, mutant.
Table 3
| Gene loci | Genotype | BMI (kg/m2) | SBP (mmHg) | DBP (mmHg) | Family history of hypertension (n) |
|---|---|---|---|---|---|
| CYP2C9*3 | WW | 23.90±3.19 | 138.77±12.32 | 84.11±6.96 | 1,029 |
| WM | 23.98±3.03 | 138.95±12.82 | 84.41±7.21 | 86 | |
| MM | 23.47±2.76 | 138.74±11.45 | 83.74±6.42 | 3 | |
| F/χ2 | 0.453 | 0.073 | 0.722 | 0.585 | |
| P | 0.636 | 0.929 | 0.486 | 0.747 | |
| ADRB1(1165G>C) | WW | 23.91±3.16 | 138.98±12.84 | 83.97±7.09 | 79 |
| WM | 23.94±3.21 | 138.96±12.43 | 84.08±7.06 | 411 | |
| MM | 23.89±3.14 | 138.65±12.25 | 84.20±6.91 | 628 | |
| F/χ2 | 0.372 | 0.765 | 0.550 | 1.253 | |
| P | 0.689 | 0.465 | 0.577 | 0.534 | |
| AGTR1(1166A>C) | WW | 23.92±3.19 | 138.73±12.26 | 84.16±6.96 | 1,017 |
| WM | 23.76±2.96 | 139.17±13.13 | 83.86±7.13 | 98 | |
| MM | 24.28±3.20 | 140.37±14.15 | 85.16±7.61 | 3 | |
| F/χ2 | 1.521 | 1.049 | 1.441 | 4.424 | |
| P | 0.219 | 0.350 | 0.237 | 0.109 | |
| CYP2D6*10 | WW | 23.90±3.27 | 138.81±12.78 | 84.13±6.88 | 234 |
| WM | 23.93±3.14 | 138.80±12.25 | 84.26±6.97 | 478 | |
| MM | 23.90±3.15 | 138.78±12.24 | 84.02±7.06 | 406 | |
| F/χ2 | 0.140 | 0.004 | 1.102 | 0.341 | |
| P | 0.870 | 0.996 | 0.332 | 0.843 | |
| ACE(I/D) | WW | 23.93±3.23 | 138.83±12.34 | 84.19±6.93 | 568 |
| WM | 23.90±3.12 | 138.76±12.31 | 84.10±7.03 | 448 | |
| MM | 23.84±3.11 | 138.71±12.67 | 84.05±7.01 | 102 | |
| F/χ2 | 0.355 | 0.053 | 0.264 | 4.140 | |
| P | 0.701 | 0.949 | 0.768 | 0.126 | |
| NPPA(2238T>C) | WW | 23.90±3.17 | 138.78±12.37 | 84.13±6.98 | 1106 |
| WM | 24.18±3.61 | 139.52±11.36 | 84.65±7.10 | 9 | |
| MM | 24.20±2.80 | 136.08±14.32 | 83.28±7.83 | 3 | |
| F/χ2 | 0.740 | 0.850 | 0.581 | 5.023 | |
| P | 0.477 | 0.427 | 0.560 | 0.081 | |
| CYP3A5*3 | WW | 23.82±3.18 | 138.74±11.86 | 83.79±6.45 | 116 |
| WM | 23.85±3.16 | 138.80±12.50 | 84.09±6.99 | 474 | |
| MM | 23.98±3.18 | 138.79±12.33 | 84.25±7.08 | 528 | |
| F/χ2 | 2.119 | 0.011 | 1.819 | 0.698 | |
| P | 0.120 | 0.989 | 0.162 | 0.705 |
WW, homozygous wildtype; WM, heterozygous mutant; MM, homozygous mutant; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Baseline characteristics
A total of 660 patients with hypertension were divided into 2 groups: the experimental group consisted of 225 males with an average age of 54.8±7.0 years and 270 females with an average age of 55.0±8.7 years; the control group consisted of 81 males with an average age of 55.3±8.9 years and 84 females with an average age of 56.1±9.5 years. A total of 16 participants were lost to follow-up due to lack of adherence and were excluded from the analysis. Groups were similar for baseline characteristics in terms of age, gender, weight, smoking, blood sugar, blood lipids, liver function, kidney function and rate of BP control, but the SBP in the experimental group was slightly higher than that in the control group (Table 4; P<0.05).
Table 4
| Experimental group (n=495) | Control group (n=165) | P | |
|---|---|---|---|
| Age (y) | 54.90±8.04 | 55.71±7.11 | 0.118 |
| Gender (M/F) | 225/270 | 81/84 | 0.225 |
| Weight (kg) | 62.56±9.24 | 64.12±10.08 | 0.143 |
| Smoking (n) | 202 | 53 | 0.134 |
| FBS (mmol/L) | 5.43±1.05 | 5.60±2.31 | 0.090 |
| TC (mmol/L) | 4.47±0.90 | 4.28±1.27 | 0.416 |
| TG (mmol/L) | 1.66±1.68 | 1.66±1.04 | 0.990 |
| LDL-C (mmol/L) | 3.02±0.99 | 2.90±1.20 | 0.572 |
| BUN (mmol/L) | 5.95±1.79 | 5.92±2.10 | 0.941 |
| Scr (μmol/L) | 83.59±23.55 | 91.58±45.62 | 0.323 |
| SUA (mmol/L) | 369.10±114.17 | 364.89±95.58 | 0.823 |
| ALT (U/L) | 32.56±0.83 | 29.88±1.53 | 0.092 |
| SBP (mmHg) | 137.92±13.12* | 133.12±9.94 | 0.027 |
| DBP (mmHg) | 90.95±11.70 | 88.35±8.55 | 0.173 |
| BP control rate (%) | 38.6±6.5 | 33.9±4.9 | 0.115 |
*, compared with the control group, P<0.05. FBS, fasting plasma glucose; TC, total cholesterol; TG, triglyceride; LDL-C, low-density lipoprotein cholesterol; BUN, serum urea nitrogen; Scr, serum creatinine; SUA, serum uric acid; ALT, alanine aminotransferase; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Analysis of clinical efficacy in pre- and post-treatment groups
In the experimental group and the control group, significant decreases in diastolic pressure (88.51±11.32 mmHg vs. 90.95±11.70 mmHg and 86.79±8.46 mmHg vs. 88.35±8.55 mmHg, respectively; P<0.01) were observed in response to adjusted antihypertensive agents. Correspondingly, in the genotype-guided group, an overall treatment effect of 3.52±11.72 mmHg in systolic pressure was observed, with pre- and postintervention values of 137.92±13.12 and 134.40±11.37 mmHg (P<0.01), respectively, with no significant difference in the reduction of systolic pressure observed in the clinical experience-guided group (P>0.05; Table 5). Pre- and post-treatment intra-group comparisons showed significant increases in the rate of BP control (47.1% vs. 38.6% and 37.5% vs. 33.9%, respectively; P<0.05; Table 6). In the experimental group, there was no significant difference in the number of patients with grade 1of BP before and after treatment, but the number of patients with grade 2 and grade 3 of BP decreased significantly after treatment (P<0.01). No significant decrease in the number of people with a different grade of hypertension was observed in the control group (Table 7).
Table 5
| Groups | n | SBP (mmHg) | DBP (mmHg) |
|---|---|---|---|
| Experimental group | |||
| Pre- | 495 | 137.92±13.12 | 90.95±11.70 |
| Post- | 484 | 134.40±11.37** | 88.51±11.32** |
| D-value | 8 | 3.52±11.72# | 2.44±11.78 |
| Control group | |||
| Pre- | 165 | 133.12±9.94 | 88.35±8.55 |
| Post- | 160 | 132.20±8.76 | 86.79±8.46** |
| D-value | 6 | 0.92±9.14 | 1.56±8.50 |
**, comparisons of intra-group pre- and post-treatment, P<0.01. #, comparisons between two groups post-treatment, P<0.01. BP, blood pressure; SBP, mean values of 24-h systolic blood pressure; DBP, mean values of 24-h diastolic blood pressure.
Table 6
| Groups | Controlled (n) | Uncontrolled (n) | Totaled (n) | Rate of BP control | Δ |
|---|---|---|---|---|---|
| Experimental group | |||||
| Pre- | 191 | 304 | 495 | 38.6% | |
| Post- | 228 | 256 | 484 | 47.1%* | 8.5%# |
| Control group | |||||
| Pre- | 56 | 109 | 165 | 33.9% | |
| Post- | 60 | 100 | 160 | 37.5%* | 3.6% |
*, comparisons of intra-group pre- and post-treatment, P<0.05. #, comparisons between groups post-treatment, P<0.01. BP, mean values of 24-h blood pressure.
Table 7
| Groups | n | Grade of BP | Pre- (n/%) | Post- (n/%) | Δ |
|---|---|---|---|---|---|
| Experimental group | |||||
| Pre- | 495 | Grade 1 | 210/42.4 | 211/43.6 | 1.2% |
| Post- | 484 | Grade 2 | 75/15.2 | 41/8.5** | 6.7%# |
| Grade 3 | 19/3.8 | 4/0.8** | 3.0%# | ||
| Control group | |||||
| Pre- | 165 | Grade 1 | 79/47.9 | 74/46.3 | 1.6% |
| Post- | 160 | Grade 2 | 25/15.2 | 22/13.8 | 1.4% |
| Grade 3 | 5/3.0 | 4/2.5 | 0.5% | ||
**, comparisons of intra-group pre- and post-treatment, P<0.01. #, comparisons between two groups post-treatment, P<0.01. BP, mean values of 24-h blood pressure.
Analysis of clinical efficacy between the 2 groups post-treatment
After 4 weeks of adjusted antihypertensive therapy, the comparisons between the 2 groups showed a significant reduction in systolic pressure (3.52±11.72 vs. 0.92±9.14 mmHg, respectively; P<0.01), whereas the reduction of diastolic pressure was not statistically significant (2.44±11.78 vs. 1.56±8.50 mmHg, respectively; P>0.05; Table 5). The increased rate of BP control and the decreased percentage of people with grade 2 and grade 3 hypertension in the experimental group was more significant than that in the control group (8.5% vs. 3.6%, 6.7% vs. 1.4% and 3.0% vs. 0.5%, respectively; P<0.01; Tables 6,7).
Discussion
Up to now, the choices of antihypertensive drugs have been based on clinical experience and relevant hypertension guidelines. A lack of expert consensus and guidelines on dosing, antihypertensive drug validity, and side effects as well as balancing clinical benefit with cost remain crucial challenges in antihypertensive therapy (11). How to prescribe the most effective antihypertensive drugs with the least amount of side effects for each hypertensive patient is a major issue for clinicians. It has been found that the heterogeneity of patients’ responses to antihypertensive drugs is, to a large extent, genetically determined (12-14). However, personalized pharmacotherapy based on genetic information has not yet been established in the field of hypertension research. To our knowledge, this was the first study to compare the value of genotype-based prescribing and clinical experience-based prescribing in antihypertensive therapy. Several hypertension studies have focused on the association between genetic polymorphic loci and the effect of different antihypertensive drugs, particularly diuretics and beta blocker (2,7). Patients with the NPPA T2238C gene mutation were more sensitive to diuretics and experienced a better therapeutic effect when treated with diuretics than other hypertensive patients (15). A genome-wide association study (GWAS) involving over 60,000 cases identified polymorphisms of ADRB1 and CYP2D6 associated with hypertension and the response to beta blockers, and the most common gene mutations of ADRB1 and CYP2D6 in the Asian population are G1165C and CYP2D6*10, respectively (16,17). The CCBs were mainly metabolized by CYP3A4 and CYP3A5 enzymes, and the polymorphic CYP3A5 genotypes have been shown to cause a different response to CCBs, such as amlodipine (18). Most data are highly conflicting such that no candidate gene shows a positive association with responses to ACEI and ARBs (19). However, genes encoding key enzymes that are involved in the metabolism, transport and targets of drugs have provided a basis for pharmacogenomics, which may also contribute to BP control in humans. For example, losartan is biotransformed into its active metabolite, EXP-3174, mediated by the metabolizing enzyme, CYP2C9. A relationship between CYP2C9 polymorphisms and the pharmacokinetics of ARBs have also been observed, although the clinical impact in terms of BP was modest (20). It has been shown that AGTR1 mediates the function of ARBs by association with G proteins that activate a phosphatidylinositol-calcium second messenger system (21). Further, ACE is a key enzyme in the enzyme/substrate cascade reaction of RAS, the polymorphism of which has been associated with the activity of ACE (3). Among the polymorphic ACE genotypes, the DD genotype has the highest level of activity, and II genotype has the lowest level of activity, with the level of activity of the ID genotype sitting in the middle. The higher activity of ACE has, the more significant antihypertensive role ACEI plays (22).
Based on the above, we performed PCR-fluorescence probe technology to detect the distribution frequency of these genetic polymorphic loci from 9,933 patients with hypertension in the Changsha County of Hunan province, and found that the mutation frequencies of CYP2C9*3, AGTR1(1166A>C), ADRB1(1165G>C), CYP2D6*10, ACE(I/D), CYP3A5*3 and NPPA(2238T>C) were 4.41%, 5.55%, 74.60%, 57.08%, 30.94%, 69.03% and 1.19%, respectively. It has been shown that the mutation of these 7 genes could decrease enzyme activity, slow down the drug metabolism and increase blood drug concentration, increasing the sensitivity to drugs (15,22-25). Our results suggested that a large group of hypertensive patients seem to exhibit more sensitivity to beta blockers and CCBs, poor sensitivity to diuretics, and normal sensitivity to ARB and ACEI. While prescribing the above drugs, appropriate dosage adjustment should be made according to the gene mutation frequency associated with responses to anti-hypertensive drugs and individual circumstances.
The 24-hour ABPM has been widely used as the most informative measurement of BP behavior because it can more accurately and comprehensively reflect a patient’s overall BP. Recently, ABPM was recommended in English, Canadian and American guidelines (9,26,27) for diagnosing hypertension, especially for identifying white-coat and masked hypertension, determining the efficacy of treatment, and assessing the long-term control of hypertension (28,29). It has been shown that clinical BP monitoring alone is inadequate for optimizing BP control because of BP variability, and ABPM should be used more routinely to confirm BP control (30). Besides, ABPM is a stronger predictor of all-cause and cardiovascular mortality than clinical BP monitoring (31). The 2017 AHA Hypertension Guidelines pointed out that the evaluation of antihypertensive efficacy should be carried out at least 2 weeks after drugs adjustment (13). Therefore, we used ABPM to detect the patients’ responses to adjusted antihypertensive drugs, and a mean 24-hour SBP of ≤130 mmHg and a 24-hour DBP of ≤80 mmHg was considered the standard.
According to the results of gene detection (as shown in Table S1), the most sensitive antihypertensive drugs with the least adverse effects were prescribed to each patient in the experimental group. If a patient’s genotype was ADRB1(1165C/C), which is highly sensitive to beta blocker, a beta-blocker was the preferred drug and a starting minimum dose was deemed appropriate. On the contrary, if a patient’s genotype is determined as ADRB1(1165G/G), which is insensitive to beta blocker, a beta-blocker should not be prescribed for antihypertensive therapy. For example, the genetic polymorphic loci associated with the effect of antihypertensive drugs of case 36 was CYP2C9*1/*1, AGTR1(1166A/A), ADRB1(1165C/C), CYP2D6*1/*1, ACE(I/D), CYP3A5*3/*3 and NPPA(2238T/T), which showed high sensitivity to CCBs and beta blocker, middle sensitivity to ARB, ACEI and diuretics. The 24-hour mean BP of this patient with a long-term use of indapamide (diuretics) as antihypertensive drug was 151/79 mmHg. Considering the antihypertensive effect of diuretics was relatively weak, therefore, indapamide was adjusted to L-amlodipine (CCB) for this patient. After 4 weeks, the 24-hour mean BP was decreased to 136/82 mmHg (up to standard). Additionally, the treatment was prescribed by the designated doctor based on the Revised Guidelines for the Prevention and Treatment of Hypertension in China 2010 in the control group. As the prescription of the control group was affected by the doctor’s clinical experience and medication preference, in order to reduce the risk of antihypertensive drugs and high test costs, the allocation ratio of the experiment group to the control group was set at 3:1. After 4 weeks of adjusted antihypertensive therapy, significant decreases in DBP were observed in both groups, while a significant difference in SBP reduction was observed in the experimental group but not in the control group. The reason for this may be that the failure to reach the standard in the control group was mainly due to high DBP, while antihypertensive drugs adjusted according to clinical experience mainly reduced DBP, hence there was no significant difference in the reduction of SBP. In addition, a significant increase in the rate of BP control was observed in both groups, and the increase of BP control rate in the experimental group was more significant than that in the control group. Only in the experimental group the number of patients with grade 2 and grade 3 of BP decreased significantly after treatment. These results indicated that genotype-guided, personalized treatment is more likely to be effective in hypertensive patients, especially in those with significantly elevated SBP.
In clinical practice, high SBP, low DBP and large pulse pressure are the characteristics of elderly patients with hypertension, and systolic pressure is independently related to cardiovascular and cerebrovascular events (32); therefore, the genotype-guided antihypertensive therapy can not only provide a new scientific basis for clinically individualized medication but also significantly reduce the side effects of antihypertensive drugs. It is also expected that genotype-guided antihypertensive therapy will reduce the incidence of hypertensive complications by reducing SBP, and ultimately reduce the economic burden on patients and society, which is worthy of application and promotion in clinical work. Notably, due to the short observation time of only 4 weeks, more clinical benefits of gene-guided individualized therapy may need to be determined by extending the observation time, expanding the sample size and using more AMBP indexes such as BP variability and circadian BP rhythm. Furthermore, the different treatments in the control group were based on age and other individual circumstances, whereas in the experimental group, prescription was exclusively based on genotype. Therefore, the analysis of curative effects should be according to age stratification in consideration of the wide age range of participants. In addition, the kinds and doses of antihypertensive drugs prescribed based on the gene phenotype and pharmacokinetic characteristics or evidence-based medicine were different between different individuals, so data on antihypertensive medication in the 2 groups were not presented. Further research will focus on the efficacy of gene-guided clinical medication in patients with very high BP, such as refractory hypertension, or on the efficacy and adverse outcomes of a specific antihypertensive drug under the guidance of genotyping for individual patients.
Acknowledgments
This work was supported by the Changsha County Government.
Funding: This work was funded by the National Natural Science Foundation of China (No. 82000339) and the Natural Science Foundation of Hunan Province (No. 2020JJ5933 and No. 2021JJ31052).
Footnote
Data Sharing Statement: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-154/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-154/coif). MFC reports that the study materials of this manuscript were provided by Changsha County Government. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of Xiangya Hospital of Central South University (No. 20170124028) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ma LY, Chen WW, Gao RL, et al. China cardiovascular diseases report 2018: an updated summary. J Geriatr Cardiol 2020;17:1-8. [PubMed]
- Melville S, Byrd JB. Personalized Medicine and the Treatment of Hypertension. Curr Hypertens Rep 2019;21:13. [Crossref] [PubMed]
- Rysz J, Franczyk B, Rysz-Górzyńska M, et al. Pharmacogenomics of Hypertension Treatment. Int J Mol Sci 2020;21:4709. [Crossref] [PubMed]
- Luizon MR, Pereira DA, Sandrim VC. Pharmacogenomics of Hypertension and Preeclampsia: Focus on Gene-Gene Interactions. Front Pharmacol 2018;9:168. [Crossref] [PubMed]
- Pinheiro DS, Santos RS, Jardim PCBV, et al. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: A genetic association study in Brazilian patients. PLoS One 2019;14:e0221248. [Crossref] [PubMed]
- Xie M, Yuan S, Zeng Y, et al. ATP2B1 gene polymorphisms rs2681472 and rs17249754 are associated with susceptibility to hypertension and blood pressure levels: A systematic review and meta-analysis. Medicine (Baltimore) 2021;100:e25530. [Crossref] [PubMed]
- Oliveira-Paula GH, Pereira SC, Tanus-Santos JE, et al. Pharmacogenomics And Hypertension: Current Insights. Pharmgenomics Pers Med 2019;12:341-59. [Crossref] [PubMed]
- Liu LSWriting Group of 2010 Chinese Guidelines for the Management of Hypertension. 2010 Chinese guidelines for the management of hypertension. Zhonghua Xin Xue Guan Bing Za Zhi 2011;39:579-615. [PubMed]
- Whelton PK, Carey RM, Aronow WS, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018;71:e13-e115. [PubMed]
- Dadlani A, Madan K, Sawhney JPS. Ambulatory blood pressure monitoring in clinical practice. Indian Heart J 2019;71:91-7. [Crossref] [PubMed]
- Eadon MT, Kanuri SH, Chapman AB. Pharmacogenomic studies of hypertension: paving the way for personalized antihypertensive treatment. Expert Rev Precis Med Drug Dev 2018;3:33-47. [Crossref] [PubMed]
- Oliveira-Paula GH, Luizon MR, Lacchini R, et al. Gene-Gene Interactions Among PRKCA, NOS3 and BDKRB2 Polymorphisms Affect the Antihypertensive Effects of Enalapril. Basic Clin Pharmacol Toxicol 2017;120:284-91. [Crossref] [PubMed]
- Zhang ZL, Li HL, Wen ZP, et al. Influence of G-protein β-Polypeptide 3 C825T Polymorphism on Antihypertensive Response to Telmisartan and Amlodipine in Chinese Patients. Chin Med J (Engl) 2016;129:8-14. [Crossref] [PubMed]
- Chen L, Xiao T, Chen L, et al. The Association of ADRB1 and CYP2D6 Polymorphisms With Antihypertensive Effects and Analysis of Their Contribution to Hypertension Risk. Am J Med Sci 2018;355:235-9. [Crossref] [PubMed]
- Lynch AI, Boerwinkle E, Davis BR, et al. Pharmacogenetic association of the NPPA T2238C genetic variant with cardiovascular disease outcomes in patients with hypertension. JAMA 2008;299:296-307. [Crossref] [PubMed]
- Wu D, Li G, Deng M, et al. Associations between ADRB1 and CYP2D6 gene polymorphisms and the response to β-blocker therapy in hypertension. J Int Med Res 2015;43:424-34. [Crossref] [PubMed]
- Thomas CD, Johnson JA. Pharmacogenetic factors affecting β-blocker metabolism and response. Expert Opin Drug Metab Toxicol 2020;16:953-64. [Crossref] [PubMed]
- Zhang YP, Zuo XC, Huang ZJ, et al. CYP3A5 polymorphism, amlodipine and hypertension. J Hum Hypertens 2014;28:145-9. [Crossref] [PubMed]
- Cooper-DeHoff RM, Johnson JA. Hypertension pharmacogenomics: in search of personalized treatment approaches. Nat Rev Nephrol 2016;12:110-22. [Crossref] [PubMed]
- Park YA, Song YB, Yee J, et al. Influence of CYP2C9 Genetic Polymorphisms on the Pharmacokinetics of Losartan and Its Active Metabolite E-3174: A Systematic Review and Meta-Analysis. J Pers Med 2021;11:617. [Crossref] [PubMed]
- Liu Y, Kong X, Jiang Y, et al. Association of AGTR1 A1166C and CYP2C9*3 Gene Polymorphisms with the Antihypertensive Effect of Valsartan. Int J Hypertens 2022;2022:7677252. [Crossref] [PubMed]
- Chan SW, Chu TTW, Ho CS, et al. Influence of CYP2D6 and CYP3A5 Polymorphisms on the Pharmacokinetics and Pharmacodynamics of Bisoprolol in Hypertensive Chinese Patients. Front Med (Lausanne) 2021;8:683498. [Crossref] [PubMed]
- Gong H, Mu L, Zhang T, et al. Association of polymorphisms of CYP11B2 gene -344C/T and ACE gene I/D with antihypertensive response to angiotensin receptor blockers in Chinese with hypertension. J Genet 2019;98:1. [Crossref] [PubMed]
- Sun Y, Liao Y, Yuan Y, et al. Influence of autoantibodies against AT1 receptor and AGTR1 polymorphisms on candesartan-based antihypertensive regimen: results from the study of optimal treatment in hypertensive patients with anti-AT1-receptor autoantibodies trial. J Am Soc Hypertens 2014;8:21-7. [Crossref] [PubMed]
- Liang H, Zhang X, Ma Z, et al. Association of CYP3A5 Gene Polymorphisms and Amlodipine-Induced Peripheral Edema in Chinese Han Patients with Essential Hypertension. Pharmgenomics Pers Med 2021;14:189-97. [Crossref] [PubMed]
- Boffa RJ, Constanti M, Floyd CN, et al. Hypertension in adults: summary of updated NICE guidance. BMJ 2019;367:l5310. [Crossref] [PubMed]
- Dionne JM, Harris KC, Benoit G, et al. Hypertension Canada's 2017 Guidelines for the Diagnosis, Assessment, Prevention, and Treatment of Pediatric Hypertension. Can J Cardiol 2017;33:577-85. [Crossref] [PubMed]
- O'Brien E, White WB, Parati G, et al. Ambulatory blood pressure monitoring in the 21st century. J Clin Hypertens (Greenwich) 2018;20:1108-11. [Crossref] [PubMed]
- Anstey DE, Muntner P, Bello NA, et al. Diagnosing Masked Hypertension Using Ambulatory Blood Pressure Monitoring, Home Blood Pressure Monitoring, or Both? Hypertension 2018;72:1200-7. [Crossref] [PubMed]
- Banegas JR, Ruilope LM, de la Sierra A, et al. High prevalence of masked uncontrolled hypertension in people with treated hypertension. Eur Heart J 2014;35:3304-12. [Crossref] [PubMed]
- Banegas JR, Ruilope LM, de la Sierra A, et al. Relationship between Clinic and Ambulatory Blood-Pressure Measurements and Mortality. N Engl J Med 2018;378:1509-20. [Crossref] [PubMed]
- Cui X, Zhao Q, Yu J, et al. Cumulative mean arterial pressure and risks of adverse cardiac and cerebrovascular events: a prospective cohort study of 53,813 adults. J Hum Hypertens 2018;32:585-93. [Crossref] [PubMed]


