
CircHSPG2 absence weakens hypoxia-induced dysfunction in cardiomyocytes by targeting the miR-25-3p/PAWR axis
Introduction
Myocardial infarction (MI) is one of the leading pathological causes of disability and mortality in cardiovascular diseases (1). The apoptosis, hypertrophy, and inflammatory response of cardiomyocytes are related to MI (2,3), which eventually results in heart failure (4,5). Cardiomyocytes are terminally differentiated cells without regenerative potential. Therefore, investigating the molecular mechanism behind hypoxia-induced dysfunction of cardiomyocytes is essential to improve the clinical treatment strategies for MI.
Circular RNAs (circRNAs) are implicated in the regulation of almost all cellular processes, and previous articles have pointed out that circRNAs play pivotal functions in the pathogenesis of multiple heart diseases, including MI, heart failure, and hypertrophy (6,7). CircRNA heparan sulfate proteoglycan 2 (circHSPG2; hsa_circ_0010729) is a newly discovered circRNA, which is related to hypoxia-induced growth of vascular endothelial cells (8). Lei et al. found that circHSPG2 silencing attenuates hypoxia-induced cardiomyocyte injuries by releasing miR-27a-3p and repressing TRAF5 expression (9). Moreover, Zhang et al. demonstrated that hypoxia exposure enhances circHSPG2 expression in cardiomyocytes, and circHSPG2 silencing suppresses hypoxia-evoked injury in cardiomyocytes through mediating miR-370-3p/TRAF6 signaling cascade (10). The potential regulatory mechanism behind the protective role of circHSPG2 in hypoxia-induced cardiomyocytes was further explored.
Accumulating evidence has demonstrated that circRNAs can regulate cell biological behaviors by acting as microRNA (miRNA) sponges (11). miRNAs play pivotal regulatory roles in cardiovascular diseases, including dilated cardiomyopathy (12), cardiac remodeling (13), and heart failure (14). Through bioinformatics analysis via Starbase database, it was found that miR-25-3p harbored the complementary sites with circHSPG2. Pan et al. demonstrated that miR-25 overexpression protects cardiomyocytes from oxidation-mediated injury by regulating mitochondrial calcium uniporter expression (15). Yao et al. found that miR-25 attenuates sepsis-evoked apoptosis of cardiomyocytes by targeting PTEN (16). Qin et al. found that miR-25 abundance is reduced in hypoxia-induced cardiomyocytes, and it facilitates cell proliferation and migration abilities and hampers cell apoptosis in cardiomyocytes by modulating Bim (17). Here, the function of miR-25-3p and its related mechanism in hypoxia-induced myocardial damage were investigated in this study.
Through bioinformatics analysis using Starbase database, it was found that pro-apoptotic WT1 regulator (PAWR) possessed the complementary sites with miR-25-3p. Chai et al. found that PAWR expression is up-regulated in hypoxia-induced cardiomyocytes, and circ_0068655 facilitates the apoptosis of cardiomyocytes by up-regulating PAWR via sponging miR-498 (18). The associated relationship of miR-25-3p and PAWR was tested.
In the current study, we first analyzed the role of circHSPG2 in MI using hypoxia-induced AC-16 cell model. Then, bioinformatics analysis was conducted to establish circHSPG2/miRNA/mRNA axis, and rescue experiments were conducted to verify the working mechanism of circHSPG2 in hypoxia-induced AC-16 cells. We aimed to identify novel targets for MI intervention and treatment.
Methods
Cell culture
Human ventricular cardiomyocytes (AC-16) (19) were purchased from Beijing Institute for Cancer Research Collection (Beijing, China). Dulbecco’s modified Eagle’s medium/F12 (#11320033; DMEM/F12; Invitrogen, Waltham, MA, USA) with the supplement of 10% fetal bovine serum (#10099141C; FBS, Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin solution (#15070063; Gibco) was utilized for the cultivation of AC-16 cells. AC-16 cells were cultured under the standard condition (5% CO2, 37 ℃). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Hypoxia treatment
AC-16 cells were cultured under the condition of 1% O2, 94% N2, and 5% CO2 for 24 h to induce hypoxia, and cells growing under normoxic condition were regarded as the control.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA samples were isolated from AC-16 cells using TRIzol reagent (#12183555; Invitrogen). The concentration and purity of RNA samples were analyzed using NanoDrop 2000 (NanoDrop Technologies, Wilmington, DE, USA). For circRNAs and mRNAs, reverse transcription was implemented using a commercial Prime-script RT reagent Kit (#RR037A; Takara, Dalian, China), and qPCR was carried out using a commercial Premix Ex Taq Kit (#RR047A; Takara). For miRNAs, complementary DNA (cDNA) was synthesized using a commercial Taqman MicroRNA Reverse Transcription Kit (#4366596; Applied Biosystems, Foster City, CA, USA), and thermal cycling reaction was carried out using a commercial Taqman Universal Master Mix II (#4440043; Applied Biosystems). qPCR reaction was conducted on 7500 Fast Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). The thermo cycling condition was listed as below: 95 ℃ for 3 min and 38 cycles of 95 ℃ for 20 sec, 60 ℃ for 30 sec, and 72 ℃ for 30 sec. All primers used in RT-qPCR were shown in Table 1. The relative fold changes were analyzed by the 2−ΔΔCt method. RT-qPCR was repeated three times with three technical repetitions each time.
Table 1
| Gene | Sequence (5'-3') |
|---|---|
| circHSPG2 | Forward: ATGCTGGGGTCTACATTTGC |
| Reverse: ACTCGTAGCGCACGTTGTAA | |
| HSPG2 | Forward: CTCGTCCCACAACGAGCAG |
| Reverse: GGTCGACTAGCTGGAACTCG | |
| miR-25-3p | Forward: GCCGAGCATTGCACTTGTCT |
| Reverse: CAGTGCAGGGTCCGAGGTAT | |
| PAWR | Forward: CGTCGGGGTCCGTACTGTAG |
| Reverse: TCGCCATATTCCCAAAGGGG | |
| GAPDH | Forward: TATGATGACATCAAGAAGGTGGT |
| Reverse: TGTAGCCAAATTCGTTGTCATAC | |
| U6 | Forward: GCTTCGGCAGCACATATACTAAAAT |
| Reverse: CGCTTCACGAATTTGCGTGTCAT |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
RNase R treatment and subcellular localization
The circular structure of circHSPG2 was tested with RNase R (Applied Biological Materials, Vancouver, Canada). RNA samples were treated with RNase R (100 µg/mL) at 37 ℃ for 20 min. RT-qPCR was implemented to analyze circHSPG2 and linear HSPG2 mRNA abundance. RT-qPCR was repeated three times with three technical repetitions each time.
The cytoplasmic and nuclear fractions of cicHSPG2 were isolated using the commercial PARISTM Kit (#AM1921; Thermo Fisher Scientific). This experiment was repeated three times.
Cell transfection
Small interfering (si)RNA against circHSPG2 (si-circHSPG2), negative control of siRNA (si-NC), overexpression plasmid of circHSPG2 (oe-circHSPG2), pLO-ciR vector (vector), mimics of miR-25-3p (miR-25-3p), miR-NC, inhibitor of miR-25-3p (anti-miR-25-3p), anti-miR-NC, PAWR overexpression plasmid (PAWR), and pcDNA3.1(+) were synthesized or constructed by Geneseed (Guangzhou, China) and GeneChem (Shanghai, China). Plasmid (1 µg), miRNA mimics (50 nM), or miRNA inhibitor (20 nM) was transfected into AC-16 cells using Lipofectamine 3000 (Invitrogen). For hypoxia and transfection co-treatment, AC-16 cells were induced by hypoxia for 24 h after transfection for 24 h.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay
A total of 5×103 AC-16 cells were seeded onto 96-well plates. The next day, AC-16 cells were incubated with 20 µL of MTT reagent (#11465007001; 5 mg/mL; Sigma, St. Louis, MO, USA) for 4 h at 37 ℃. Then, 100 µL of dimethyl sulfoxide (#11465007001; DMSO, Sigma) was pipetted to the wells to dissolve the formazan products, and the plates were gently shaken for 10 min. The optical density was examined at the wavelength of 570 nm under a microplate reader (Bio-Rad). MTT assay was repeated three times with six technical repetitions each time.
5-Ethynyl-2'-deoxyuridine (EdU) assay
AC-16 cells in 96-well plates were incubated with 50 µM EdU (#C10310-1; RiboBio, Guangzhou, China) for 1 h followed by immobilization with 100 µL of 4% paraformaldehyde (Sangon Biotech, Shanghai, China) at 25 ℃ for 30 min. Cell nucleus was marked with 100 ng/mL 4', 6-diamidino-2-phenylindole (#MBD0015; DAPI; Sigma) for 10 min. Five random fields were captured under a Nikon fluorescence microscope (Eclipse Ti2‑U; Nikon) and the positive rate was analyzed. EdU assay was repeated three times with three technical repetitions each time.
Colony formation assay
AC-16 cells were seeded onto 12-well plates at 200 cells/well and were continued to culture for 12 d. The culture supernatant was replenished every 4 d. The colonies were immobilized with 4% paraformaldehyde (#E672002-0500; Sangon Biotech) for 15 min at room temperature, and were then stained with 0.5% crystal violet for 10 min at room temperature. Colony number (with more than 50 cells) was analyzed under an optical microscope (Olympus, Tokyo, Japan). Colony formation assay was repeated three times with three technical repetitions each time.
Flow cytometry (FCM) analysis
Cell apoptosis was analyzed by fluorescein isothiocyanate (FITC) and propidium iodide (PI) double-staining using the commercial Apoptosis Detection Kit (#40302ES20; Qcbio Science & Technologies, Shanghai, China). AC-16 cells were dispersed in 100 µL of binding buffer, and then cells were incubated with 10 µL of Annexin V-FITC and 10 µL of PI for 15 min at room temperature in the dark. Cells were loaded onto the flow cytometer (Beckman Coulter, Fullerton, CA, USA) for the analysis of apoptosis rate. FCM analysis was repeated three times with three technical repetitions each time.
Western blot assay
Proteins in AC-16 cells were isolated with the radio-immunoprecipitation assay (RIPA) lysis buffer (Sigma). Protein samples (35 µg) were loaded onto 10% separating gel and blotted onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). After blocking with 5 mL 5% skimmed milk for 1 h at room temperature, the primary antibodies were incubated with the membrane overnight at 4 ℃. The next day, the membrane was incubated with the secondary antibody for 2 h at room temperature. Protein blots were analyzed using the enhanced chemiluminescence (ECL) kit (Pierce, Waltham, MA, USA). The intensities of protein bands were quantified using the Image Lab analysis software (National Institutes of Health, Bethesda, MD, USA). The primary antibodies were all purchased from Abcam (Cambridge, MA, USA), including anti-B cell leukemia/lymphoma 2 (anti-Bcl-2; 1/1000; ab32124), anti-Bcl-2 associated X, apoptosis regulator (anti-Bax; 1/1000; ab32503), anti-PAWR (ab92590; 1/10000), and anti-GAPDH (ab8245; 1/5000). Western blot assay was repeated three times.
Lactate dehydrogenase (LDH) assay
Cell death was assessed by quantifying the level of LDH. The release of LDH in the culture supernatant was analyzed using the LDH cytotoxicity assay kit (#C0017; Beyotime, Haimen, China) according to the manufacturer’s instructions. In brief, a total of 5×103 AC-16 cells were seeded onto 96-well plates. Then, 120 µL of culture supernatant was incubated with 60 µL of LDH test working reagent for 30 min in the dark. The absorbance was detected at the wavelength of 490 nm under a microplate reader (Bio-Rad). LDH assay was repeated three times with six technical repetitions each time.
Bioinformatics analysis
The circHSPG2-miRNA interactions and miR-25-3p-mRNA interactions was established by Starbase database (http://starbase.sysu.edu.cn).
Dual-luciferase reporter assay
The fragment of circHSPG2 or PAWR 3’ untranslated region (3’UTR), including the wild-type or mutant binding sites with miR-25-3p, was inserted into pmirGLO plasmid (Promega, Madison, WI, USA) to generate WT/MUT-circHSPG2 and WT/MUT-PAWR 3’UTR. AC-16 cells were seeded onto 24-well plates at 4×104 cells/well. The luciferase plasmids (100 ng) were transfected into AC-16 cells with miR-25-3p (50 nM) or miR-NC (50 nM) using Lipofectamine 3000 (Invitrogen). After transfection for 48 h, the luciferase intensities were examined using the commercial dual-luciferase reporter assay system kit (Promega). The intensities of fluorescence were determined using a GloMax 20/20 luminometer (Promega). Firefly luciferase intensity was normalized to Renilla luciferase activity. Dual-luciferase reporter assay was repeated three times.
RNA immunoprecipitation (RIP) assay
AC-16 cells were plated onto the 6-well plates at 2×105 cells/well. AC-16 cells transfected with miR-NC (50 nM) or miR-25-3p (50 nM) were disrupted with the RIP buffer for 5 min on the ice, and then cell lysates were mixed with magnetic beads conjugated with Argonaute 2 (Ago2) antibody (ab186733; 1: 50; Abcam) or immunoglobulin G (IgG) antibody (ab172730; 1: 100; Abcam) for 3 h at 4 ℃. The beads were washed with 500 µL RIP wash buffer twice and were incubated with RIP Immunoprecipitation buffer. The mixture was then centrifuged at 14,000 rpm for 10 min at 4 ℃. RNA was extracted with TRIzol reagent (Invitrogen), and the immune-precipitated RNAs were examined by RT-qPCR. RIP assay was repeated three times.
Statistical analysis
Data were processed by GraphPad Prism 7 software (GraphPad) and were represented as mean ± standard deviation (SD). The differences were assessed by Student’s t-test (two groups) or one-way analysis of variance (ANOVA) followed by Tukey’s test (three or more groups). P<0.05 was considered statistically significant.
Results
Hypoxia treatment up-regulates circHSPG2 expression in AC-16 cells
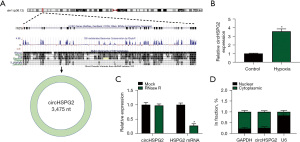
The chromosomal localization of circHSPG2 (3475 nt) was shown in Figure 1A. We found that circHSPG2 was significantly up-regulated after hypoxia exposure for 24 h in AC-16 cells (Figure 1B). CircHSPG2 was resistant to the degradation of RNase R, while RNase R treatment markedly reduced the level of its linear counterpart HSPG2 (Figure 1C), suggesting that circHSPG2 was indeed a circular transcript. Before exploring the biological function of circHSPG2, we first analyzed the subcellular localization of circHSPG2. With GAPDH or U6 as cytoplasmic or nuclear indicator, it was found that circHSPG2 was majorly distributed in the cytoplasmic fraction of AC-16 cells (Figure 1D), suggesting that circHSPG2 might function in post-transcriptional level.
CircHSPG2 absence alleviates hypoxia-induced dysfunction in AC-16 cells
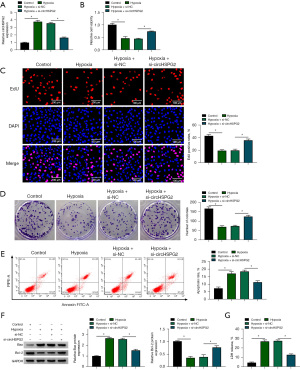
To explore the biological significance behind the abnormal up-regulation of circHSPG2 in hypoxia-induced AC-16 cells, we performed loss-of-function experiments. Hypoxia-induced up-regulation of circHSPG2 in AC-16 cells was neutralized by si-circHSPG2 (Figure 2A). It was observed that hypoxia exposure reduced the viability of AC-16 cells, and this suppressive effect was partly diminished by si-circHSPG2 (Figure 2B). Hypoxia treatment suppressed the proliferation of AC-16 cells, evidenced by the reduced percentage of EdU+ cells and number of colonies (Figure 2C,2D). The addition of si-circHSPG2 partly restored the proliferation ability in hypoxia-induced AC-16 cells (Figure 2C,2D). Hypoxia-induced the apoptosis of AC-16 cells, which was alleviated by the introduction of si-circHSPG2 (Figure 2E). Two apoptosis-associated proteins, including pro-apoptotic protein Bax and anti-apoptotic protein Bcl-2, were measured by Western blot assay. Hypoxia-induced up-regulation of Bax and down-regulation of Bcl-2 were largely reversed by si-circHSPG2 (Figure 2F). Combined with the results in Figure 2E, these data demonstrated that hypoxia-induced apoptosis in AC-16 cells was partly dependent on the up-regulation of circHSPG2. LDH assay demonstrated that hypoxia-induced cell death was largely attenuated by the silence of circHSPG2 in AC-16 cells (Figure 2G). Taken together, hypoxia-induced dysfunction of AC-16 cells was partly based on the up-regulation of circHSPG2.
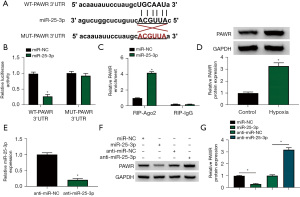
CircHSPG2 negatively regulates miR-25-3p expression by binding to it
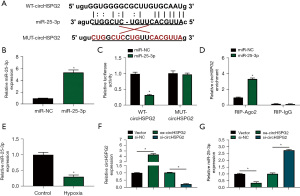
Bioinformatics software Starbase was utilized to predict the miRNA targets of circHSPG2, and miR-25-3p was predicted as one of the targets of circHSPG2. The putative binding sequence between circHSPG2 and miR-25-3p was shown in Figure 3A. RT-qPCR assay confirmed that miR-25-3p mimic (miR-25-3p) was effective in up-regulating miR-25-3p level in AC-16 cells (Figure 3B). Dual-luciferase reporter assay and RIP assay were conducted to verify whether circHSPG2 can bind to miR-25-3p. The luciferase activity was significantly reduced in WT-circHSPG2 group with the overexpression of miR-25-3p, while the luciferase activity was unaffected in MUT-circHSPG2 group with the transfection of miR-NC or miR-25-3p (Figure 3C), demonstrating that circHSPG2 directly interacted with miR-25-3p. RIP assay revealed that circHSPG2 can bind to exogenous miR-25-3p in RNA-induced silencing complex (RISC) when using Ago2 antibody (Figure 3D). Hypoxia treatment reduced the expression of miR-25-3p in AC-16 cells (Figure 3E). Then, we analyzed whether circHSPG2 can regulate the expression of miR-25-3p in AC-16 cells. RT-qPCR combined with divergent primers verified the transfection efficiency of circHSPG2 overexpression plasmid (oe-circHSPG2) and si-circHSPG2 in AC-16 cells (Figure 3F). CircHSPG2 overexpression notably reduced the expression of miR-25-3p, and circHSPG2 absence markedly up-regulated miR-25-3p level in AC-16 cells (Figure 3G). These results together demonstrated that miR-25-3p was a target of circHSPG2, and it was negatively regulated by circHSPG2 in AC-16 cells.
CircHSPG2 silencing-mediated protective effects in hypoxia-induced AC-16 cells are partly overturned by anti-miR-25-3p
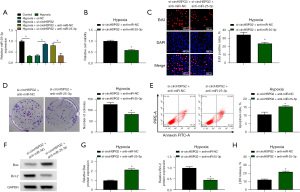
Considering that circHSPG2 absence up-regulated the expression of miR-25-3p, we co-transfected hypoxia-induced AC-16 cells with si-circHSPG2 and anti-miR-25-3p to explore whether circHSPG2 silencing-induced effects were partly dependent on the up-regulation of miR-25-3p. The addition of anti-miR-25-3p reduced the expression of miR-25-3p in AC-16 cells (Figure 4A). Cell viability was decreased by the addition of anti-miR-25-3p in AC-16 cells (Figure 4B). The introduction of anti-miR-25-3p suppressed the proliferation of AC-16 cells (Figure 4C,4D). The addition of anti-miR-25-3p triggered the apoptosis of AC-16 cells (Figure 4E). The protein expression of Bax was up-regulated while the protein level of Bcl-2 was reduced by silencing miR-25-3p (Figure 4F,4G). The addition of anti-miR-25-3p induced cell death (Figure 4H). Overall, circHSPG2 silencing-mediated protective effects in hypoxia-induced AC-16 cells were largely based on the up-regulation of miR-25-3p.
miR-25-3p binds to the 3’UTR of PAWR in AC-16 cells
The possible mRNA targets of miR-25-3p were predicted by Starbase database, and the putative binding sequence between miR-25-3p and PAWR was shown in Figure 5A. The overexpression of miR-25-3p significantly reduced the luciferase activity of WT-PAWR 3'UTR rather than MUT-PAWR 3'UTR (Figure 5B), suggesting that miR-25-3p directly interacted with the 3'UTR of PAWR. RIP assay revealed that there was spatial interaction between miR-25-3p and PAWR in RISC (Figure 5C). The expression of PAWR protein was markedly up-regulated in AC-16 cells upon hypoxia treatment (Figure 5D) and si-PAWR introduction repressed PAWR protein levels in AC-16 cells with hypoxia treatment (Figure S1A). Also, PAWR silencing weakened hypoxia-mediated effects on AC-16 cell proliferation, apoptosis, and cytotoxicity (Figure S1B-S1G). High transfection efficiency of anti-miR-25-3p was verified by RT-qPCR in AC-16 cells (Figure 5E). miR-25-3p overexpression reduced the protein expression of PAWR, and miR-25-3p knockdown up-regulated the level of PAWR protein in AC-16 cells (Figure 5F,5G). Taken together, miR-25-3p negatively regulated PAWR expression by interacting with its 3'UTR in AC-16 cells.
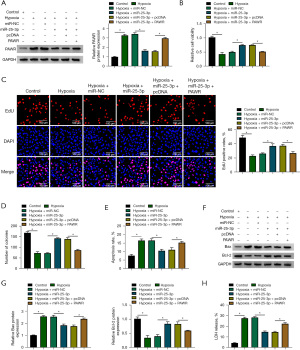
miR-25-3p overexpression protects AC-16 cells from hypoxia-induced dysfunction partly by down-regulating PAWR
Rescue experiments were conducted to analyze whether miR-25-3p regulated the biological phenotypes of AC-16 cells by targeting PAWR. miR-25-3p overexpression reduced the protein expression of PAWR, and the protein level of PAWR was largely recovered by the addition of PAWR plasmid (Figure 6A). miR-25-3p overexpression protected AC-16 cells from hypoxia-induced dysfunction in AC-16 cells (Figure 6B-6H). Furthermore, the addition of PAWR plasmid reduced the viability and suppressed the proliferation of AC-16 cells (Figure 6B-6D). PAWR overexpression also induced the apoptosis of AC-16 cells (Figure 6E). The protein level of Bax was up-regulated while the protein level of Bcl-2 was decreased by the introduction of PAWR plasmid (Figure 6F,6G). LDH assay displayed that cell death was induced by the addition of PAWR plasmid (Figure 6H). Collectively, miR-25-3p exerted a protective role in hypoxia-induced AC-16 cells partly by down-regulating PAWR.
CircHSPG2 acts as a positive regulator of PAWR by sequestering miR-25-3p in AC-16 cells
CircHSPG2 absence reduced the protein expression of PAWR, and the protein level of PAWR was largely rescued by the addition of anti-miR-25-3p in AC-16 cells (Figure 7A). CircHSPG2 overexpression increased the protein expression of PAWR, and addition of miR-25-3p decreased the protein level of PAWR in AC-16 cells (Figure 7B). These results suggested that circHSPG2 positively regulated PAWR expression by sponging miR-25-3p in AC-16 cells.

Discussion
Due to the high stability, abundant expression, and tissue-specific expression pattern, circRNAs are considered as promising bio-markers for human diseases (20). Nevertheless, the biological roles of circRNAs in cardiac pathophysiology remain largely unknown. Several articles reported that circRNAs are dysregulated in multiple cardiovascular diseases. For instance, circ_000203 is reported to be up-regulated in mouse model and cell model of cardiac hypertrophy, and circ_000203 facilitates cardiac hypertrophy progression by targeting miR-26b-5p/miR-140-3p-Gata4 axis (21). CircTLK1 is reported to be up-regulated in myocardial ischemia/reperfusion injury mouse model, and it aggravates ischemia/reperfusion injury through mediating miR-214/RIPK1 signaling (22). As for circHSPG2, previous studies found that circHSPG2 absence exerts a protective role in cardiomyocytes upon the stimulation of hypoxia or oxygen-glucose deprivation (9,23,24). We found that hypoxia exposure enhanced the expression of circHSPG2 in cardiomyocytes. Hypoxia treatment inhibited the viability and proliferation and induced the apoptosis of cardiomyocytes, and circHSPG2 silencing using si-circHSPG2 attenuated hypoxia-induced dysfunction in cardiomyocytes, suggesting that the up-regulation of circHSPG2 was important for hypoxia-induced dysfunction in cardiomyocytes. A previous report showed that hyperoxia gradually increases cellular inflammation and cytotoxicity (25), and whether hyperoxia affects the level of circHSPG2 in cardiomyocytes can be further explored in the future.
Accumulating evidence have verified that circRNAs can sequester miRNAs to regulate the expression and biological roles of miRNAs (11,26). Through bioinformatics analysis using Starbase software and experimental verification, the associated relationship between circHSPG2 and miR-25-3p was validated. Hypoxia stimulation reduced the level of miR-25-3p, and miR-25-3p was reversely modulated by circHSPG2 in AC-16 cells. miR-25-3p is identified to be a pro-tumor factor in multiple cancers (27-30). As for hypoxia-induced myocardial damage, Qin et al. found that miR-25 abundance is reduced in hypoxia-induced cardiomyocytes, and it facilitates the proliferation and migration capacities and reduces the apoptotic rate of cardiomyocytes (17). Two previous studies reported that miR-25 protects cardiomyocytes from oxidation or sepsis-induced injury by targeting mitochondrial calcium uniporter (15) or PTEN (16), respectively. Consistently, we found that miR-25-3p overexpression largely restored the viability and proliferation and suppressed the apoptosis of hypoxia-treated AC-16 cells. To investigate whether circHSPG2 functioned by targeting miR-25-3p in hypoxia-induced AC-16 cells, rescue experiments were implemented. The results presented that circHSPG2 absence protected AC-16 cells from hypoxia-induced dysfunction partly by up-regulating miR-25-3p, which further demonstrated the protective role of miR-25-3p in AC-16 cells upon hypoxia stimulation.
The interaction between miR-25-3p and PAWR was identified in AC-16 cells. It was observed that hypoxia stimulation up-regulated the protein expression of PAWR in AC-16 cells. PAWR was reversely modulated by miR-25-3p in AC-16 cells. Previous articles reported that PAWR acts as an anti-tumor factor in multiple cancers by inhibiting cell proliferation and inducing cell apoptosis (31-33). Moreover, it is reported that circ_0068655 facilitates the apoptosis of cardiomyocytes by binding to miR-498 to up-regulate the expression of PAWR (18). We found that miR-25-3p overexpression-mediated protective effects in hypoxia-induced AC-16 cells were largely reversed by the addition of PAWR plasmid, indicating that miR-25-3p protected AC-16 cells from hypoxia-induced dysfunction partly by down-regulating PAWR. Finally, it was found that circHSPG2 acted as a positive regulator of PAWR by acting as miR-25-3p sponge in AC-16 cells. FBXWJ is reported as another target of miR-25 (34), and whether FBXWJ is regulated by the circHSPG2/miR-25-3p axis can be explored in the future.
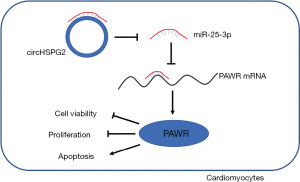
In summary, the data in this study together presented that circHSPG2 absence protected AC-16 cells from hypoxia-induced injury through mediating miR-25-3p/PAWR axis (Figure 8). Our results suggested that circHSPG2 knockdown might be a promising therapeutic strategy to alleviate hypoxia-induced myocardial damage. In future, animal experiment is needed to analyze the effect, side effect, and specificity of circHSPG2 knockdown therapy in myocardial infarction treatment.
Acknowledgments
Thanks for all participants involved in this study.
Funding: This study was supported by Research Project of health industry in Hainan Province (No. 20A200060).
Footnote
Data Sharing Statement: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-197/dss
Peer Review File: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-197/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-197/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- George TA, Hsu CC, Meeson A, et al. Nanocarrier-Based Targeted Therapies for Myocardial Infarction. Pharmaceutics 2022;14:930. [Crossref] [PubMed]
- Frangogiannis NG. Pathophysiology of Myocardial Infarction. Compr Physiol 2015;5:1841-75. [Crossref] [PubMed]
- He L, Chen X. Cardiomyocyte Induction and Regeneration for Myocardial Infarction Treatment: Cell Sources and Administration Strategies. Adv Healthc Mater 2020;9:e2001175. [Crossref] [PubMed]
- Bahit MC, Kochar A, Granger CB. Post-Myocardial Infarction Heart Failure. JACC Heart Fail 2018;6:179-86. [Crossref] [PubMed]
- Jenča D, Melenovský V, Stehlik J, et al. Heart failure after myocardial infarction: incidence and predictors. ESC Heart Fail 2021;8:222-37. [Crossref] [PubMed]
- Li M, Ding W, Sun T, et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J 2018;285:220-32. [Crossref] [PubMed]
- Fan X, Weng X, Zhao Y, et al. Circular RNAs in Cardiovascular Disease: An Overview. Biomed Res Int 2017;2017:5135781. [Crossref] [PubMed]
- Dang RY, Liu FL, Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys Res Commun 2017;490:104-10. [Crossref] [PubMed]
- Lei D, Wang Y, Zhang L, et al. Circ_0010729 regulates hypoxia-induced cardiomyocyte injuries by activating TRAF5 via sponging miR-27a-3p. Life Sci 2020;262:118511. [Crossref] [PubMed]
- Zhang J, Gao C, Zhang J, et al. Circ_0010729 knockdown protects cardiomyocytes against hypoxic dysfunction via miR-370-3p/TRAF6 axis. EXCLI J 2020;19:1520-32. [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Jiao M, You HZ, Yang XY, et al. Circulating microRNA signature for the diagnosis of childhood dilated cardiomyopathy. Sci Rep 2018;8:724. [Crossref] [PubMed]
- Batkai S, Bär C, Thum T. MicroRNAs in right ventricular remodelling. Cardiovasc Res 2017;113:1433-40. [Crossref] [PubMed]
- Wu T, Chen Y, Du Y, et al. Serum Exosomal MiR-92b-5p as a Potential Biomarker for Acute Heart Failure Caused by Dilated Cardiomyopathy. Cell Physiol Biochem 2018;46:1939-50. [Crossref] [PubMed]
- Pan L, Huang BJ, Ma XE, et al. MiR-25 protects cardiomyocytes against oxidative damage by targeting the mitochondrial calcium uniporter. Int J Mol Sci 2015;16:5420-33. [Crossref] [PubMed]
- Yao Y, Sun F, Lei M. miR-25 inhibits sepsis-induced cardiomyocyte apoptosis by targetting PTEN. Biosci Rep 2018;38:BSR20171511. [Crossref] [PubMed]
- Qin X, Gao S, Yang Y, et al. microRNA-25 promotes cardiomyocytes proliferation and migration via targeting Bim. J Cell Physiol 2019;234:22103-15. [Crossref] [PubMed]
- Chai Q, Zheng M, Wang L, et al. Circ_0068655 Promotes Cardiomyocyte Apoptosis via miR-498/PAWR Axis. Tissue Eng Regen Med 2020;17:659-70. [Crossref] [PubMed]
- Davidson MM, Nesti C, Palenzuela L, et al. Novel cell lines derived from adult human ventricular cardiomyocytes. J Mol Cell Cardiol 2005;39:133-47. [Crossref] [PubMed]
- Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018;34:267-74. [Crossref] [PubMed]
- Li H, Xu JD, Fang XH, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res 2020;116:1323-34. [Crossref] [PubMed]
- Song YF, Zhao L, Wang BC, et al. The circular RNA TLK1 exacerbates myocardial ischemia/reperfusion injury via targeting miR-214/RIPK1 through TNF signaling pathway. Free Radic Biol Med 2020;155:69-80. [Crossref] [PubMed]
- Jin Q, Chen Y. Silencing circular RNA circ_0010729 protects human cardiomyocytes from oxygen-glucose deprivation-induced injury by up-regulating microRNA-145-5p. Mol Cell Biochem 2019;462:185-94. [Crossref] [PubMed]
- Wang Y, Wu C, Zhang Y, et al. Screening for differentially expressed circRNA between Kashin-Beck disease and osteoarthritis patients based on circRNA chips. Clin Chim Acta 2020;501:92-101. [Crossref] [PubMed]
- Hafner C, Wu J, Tiboldi A, et al. Hyperoxia Induces Inflammation and Cytotoxicity in Human Adult Cardiac Myocytes. Shock 2017;47:436-44. [Crossref] [PubMed]
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016;238:42-51. [Crossref] [PubMed]
- Chen H, Pan H, Qian Y, et al. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer 2018;17:4. [Crossref] [PubMed]
- Ning L, Zhang M, Zhu Q, et al. miR-25-3p inhibition impairs tumorigenesis and invasion in gastric cancer cells in vitro and in vivo. Bioengineered 2020;11:81-90. [Crossref] [PubMed]
- Wan W, Wan W, Long Y, et al. MiR-25-3p promotes malignant phenotypes of retinoblastoma by regulating PTEN/Akt pathway. Biomed Pharmacother 2019;118:109111. [Crossref] [PubMed]
- Rao HC, Wu ZK, Wei SD, et al. MiR-25-3p Serves as an Oncogenic MicroRNA by Downregulating the Expression of Merlin in Osteosarcoma. Cancer Manag Res 2020;12:8989-9001. [Crossref] [PubMed]
- Song R, Li Y, Hao W, et al. Circular RNA MTO1 inhibits gastric cancer progression by elevating PAWR via sponging miR-199a-3p. Cell Cycle 2020;19:3127-39. [Crossref] [PubMed]
- Yang K, Shen J, Chen SW, et al. Upregulation of PAWR by small activating RNAs induces cell apoptosis in human prostate cancer cells. Oncol Rep 2016;35:2487-93. [Crossref] [PubMed]
- Pereira MC, de Bessa-Garcia SA, Burikhanov R, et al. Prostate apoptosis response-4 is involved in the apoptosis response to docetaxel in MCF-7 breast cancer cells. Int J Oncol 2013;43:531-8. [Crossref] [PubMed]
- Wang B, Xu M, Li M, et al. miR-25 Promotes Cardiomyocyte Proliferation by Targeting FBXW7. Mol Ther Nucleic Acids 2020;19:1299-308. [Crossref] [PubMed]


