
Nomogram developed with APOA5 genetic variant rs662799 and clinical characteristics predicting risk of essential hypertension in a Chinese population
Highlight box
Key findings
• The G allele of the rs662799 variant was an independent risk factor for essential hypertension (EHT). The nomogram based on rs662799 G allele, age, body mass index, smoking, diabetes, education, low-density lipoprotein cholesterol, and triglyceride metabolism in our study can provide a visual assessment of the risk of EHT in the Chinese population.
What is known and what is new?
• Hypertension is a leading cause of cardiovascular disease and of premature mortality worldwide. Genetic polymorphisms of apolipoprotein A5 (APOA5) is linked to an elevated risk of atherosclerosis, metabolic syndrome, stroke, and coronary artery disease.
• The rs662799 variant of the APOA5 gene was significantly associated with EHT.
What is the implication, and what should change now?
• A model composed of APOA5 gene polymorphism and clinical characteristics can effectively predict the risk of hypertension in a Chinese population, providing new insights for the diagnosis and treatment of hypertension.
Introduction
Hypertension is a leading cause of cardiovascular disease and premature mortality worldwide (1), and essential hypertension (EHT) is the most prevalent type, affecting 85% of patients with hypertension (2). The global prevalence of hypertension is increasing due to both an aging population and the rise of risk factors such as an unhealthy diet (e.g., high-sodium and low-potassium intake) and physical inactivity (3). However, emerging evidence indicates that genetic factors also play a pivotal role in the development and progression of hypertension (4). A recent genome-wide association study (GWAS) identified 1,477 common single nucleotide polymorphisms (SNPs) that are significantly associated with various blood pressure phenotypes (5).
Apolipoprotein A5 (ApoA5) is synthesized and secreted by the liver (6). In the early 1900s, Pennacchio et al. (7) identified a novel apolipoprotein gene located on human chromosome 11q23 by comparing genomic DNA sequences between humans and mice. APOA5 is closely related to lipid metabolism, especially the regulation of triglyceride (TG) metabolism. APOA5 protein reduces plasma TGs by inhibiting very-low-density lipoprotein (VLDL)-TG synthesis and promoting its hydrolysis (8,9). Several studies have demonstrated a clear association between APOA5 and the development of metabolic syndrome (10,11) and cardiovascular events (12).
Moreover, recent research has also indicated the involvement of the rs662799 polymorphism of APOA5 gene in influencing the level of TG and high-density lipoprotein cholesterol (HDL-C), and thus the risk of myocardial infarction (13). Although these factors have been demonstrated to be closely associated with the development of EHT, the relationship between the rs662799 polymorphism of the APOA5 gene and EHT remains unclear. In this study, we aimed to comprehensively examine the potential correlation between the rs662799 polymorphism and the susceptibility to EHT in a Chinese population using a systematic analysis. In order to identify patients at risk of hypertension, a nomogram (14) model was developed and internally validated based on independent risk factors. We present this article in accordance with the STROBE reporting checklist to ensure comprehensive and transparent reporting of our research methods and findings (available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-289/rc).
Methods
Subjects
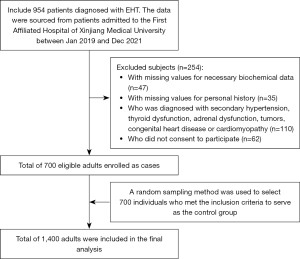
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and received ethical approval from the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University (No. K202309-08). Informed consent was obtained from all research participants. The study includes 1,400 patients (496 females and 904 males) admitted to the First Affiliated Hospital of Xinjiang Medical University between Jan 2019 and Dec 2021: 700 patients diagnosed with EHT and 700 controls. The confirmation of elevated blood pressure for defining EHT was conducted by qualified healthcare professionals. These professionals were experienced doctors specialized in the field of hypertension or trained medical staff under the supervision of doctors. According to the “Chinese Hypertension Prevention and Treatment Guidelines (2018)”, EHT in our study was defined as a systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg without the use of antihypertensive medication. To ensure the accuracy of the measurements, we followed the guideline-recommended practice of taking at least three separate measurements on different days. The choice of different days was made to account for potential day-to-day variability in blood pressure readings. The specific time interval between the measurements was determined based on the clinical judgment of the healthcare professionals involved in the study, taking into consideration the stability of the participants’ blood pressure levels. EHT was also defined as those patients with a confirmed diagnosis of hypertension and undergoing treatment with antihypertensive medication. The study excluded individuals with secondary hypertension, including conditions such as primary aldosteronism and pheochromocytoma, as well as those with thyroid dysfunction or adrenal dysfunction. A total of 700 individuals were enrolled as control participants, and any individuals with other types hypertension, tumors, congenital heart disease, or cardiomyopathy or those who declined to provide consent were excluded from the control group. According to the “Chinese Type 2 Diabetes Prevention and Treatment Guidelines (2020)”, individuals with fasting plasma glucose (FPG) levels ≥7.0 mmol/L, plasma glucose levels ≥11.1 mmol/L after a 2-hour load, with a documented history of hypoglycemic treatment were considered to have type 2 diabetes mellitus (T2DM). All the participants completed a health and lifestyle questionnaire, which included an item on smoking and alcohol intake. Figure 1 shows the inclusion and exclusion criteria used to select the study subjects.
Biochemical analysis
To ensure data accuracy and minimize the influence of other factors, we use the analysis results of the first blood sample collected during the participants’ hospital admission examination during Jan 2019 and Dec 2021. Blood samples were collected from the antecubital vein after a 12-hour overnight fasting period with vacutainer tubes containing ethylene diamine tetraacetic acid (EDTA) as an anticoagulant. After centrifugation, the blood samples were subsequently stored at a temperature of −20 ℃. The serum concentrations of various lipid parameters including HDL-C, total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), apolipoprotein A1 (ApoA1), TG, lipoprotein A [Lp(a)], apolipoprotein B (ApoB), blood uric acid (UA), blood potassium, and glucose were measured based on enzymatic methods. All biochemical measurements were performed at the Biochemistry Laboratory of the First Affiliated Hospital of Xinjiang Medical University.
Genotyping
The genomic DNA was extracted from peripheral blood leukocytes using the standard phenol-chloroform method. Subsequently, the DNA samples were carefully stored at a temperature of −80 ℃ to ensure their integrity and suitability for further analysis and experimentation. SNP analysis of APOA5 (rs662799) was conducted, with a minor allele frequency (MAF) threshold of ≥0.05 and a linkage disequilibrium pattern of r2≥0.8. This analysis was performed using Haploview software (version 4.2) and data from the International HapMap Project website phase I and II database (http://www.hapmap.org). The genotyping of the SNP was carried out using the improved multiplex ligation detection reaction (iMLDR) method. The genotyping process was conducted in a blinded manner, meaning that the individuals performing the analysis were unaware of any clinical information pertaining to the participants. In order to ensure the accuracy and reliability of the results, approximately 10% of the samples were duplicated as a quality control measurement.
SNP information
Candidate SNP sites of the APOA5 gene were acquired from an online source (http://www.ncbi.nlm.nih.gov/SNP). The following information was obtained for rs662799: consequence: intron variant; position: chr11: 116663707. The primers were arranged in the following order:
- Identification primer 1: CCCAGGAACTGGAGCGAAACTG;
- Identification primer 2: CCCAGGAACTGGAGCGAAACTA;
- Universal primer: AGATTTGCCCCATGAGGAAAAG.
Statistical analysis
Continuous parametric variables are reported as the mean ± standard deviation (SD). The Student’s t-test was used to compare quantitative traits that followed a normal distribution. Categoric variables were compared using chi-squared tests. To determine the differences in genotype and allele frequencies between two groups, the Hardy-Weinberg equilibrium (HWE) was assessed with the chi-squared test. Distributions of genotypes and alleles between two groups were also analyzed with a chi-squared test. Multivariate logistic regression analysis was performed to calculate odds ratios (ORs) and 95% confidence intervals (CIs) in order to evaluate the impact of the major risk factors.
Significant risk factors in multivariate analysis were selected to construct a prediction nomogram. In the nomogram, all predictive variables were projected to obtain the matching points on the ruler at the top then summed to get the total score, with a corresponding prediction probability below. The higher the total score, the greater the experiencing of EHT. The area under the curve (AUC) derived from the receiver operating characteristic (ROC) curve was calculated to evaluate discrimination accuracy of the nomogram. The optimal threshold was calculated using Youden index. Furthermore, we assessed the calibration ability of the model using the Hosmer-Lemeshow test, a commonly used statistical method to compare the agreement between predicted and observed outcomes. The test involves dividing the sample into groups based on predicted probabilities and calculating the difference between observed and expected outcomes in each group. A statistical test is then conducted to determine if there is a significant difference between the predicted calibration curve and the ideal curve. All of the statistical significance in our study was determined at an α level of 0.05 and all of the P value tests conducted in this study were two-sided. SPSS 21.0 (IBM Corp., Armonk, NY, USA) was used to conduct statistical analyses, and the nomogram was developed with R language (The R Foundation for Statistical Computing, Vienna, Austria).
Results
This study enrolled a total of 700 cases with EHT (450 males and 250 females; mean age 57.27±9.51 years) and 700 controls (454 males and 246 females; mean age 54.74±9.65 years). The baseline characteristics of the two groups are summarized in Table 1. Significant differences between cases and controls were found in the following characteristics: age (P<0.001), body mass index (BMI) (P<0.001), smoking (P=0.004), diabetes (P<0.001), education level (P=0.003), SBP (P<0.001), DBP (P<0.001), UA (P<0.001), FBG (P=0.026), TG (P<0.001), HDL-C (P=0.004), and LDL-C (P=0.001). No significant differences were observed in gender, drinking, pulse rate (PR), TC, ApoA1, ApoB, or Lp(a).
Table 1
| Characteristics | EHT (n=700) | Control (n=700) | χ²/t | P value |
|---|---|---|---|---|
| Male (%) | 64.3 | 64.9 | 0.050 | 0.867 |
| Age (years) | 57.27±9.51 | 54.74±9.65 | −4.943 | <0.001* |
| BMI (kg/m2) | 27.57±4.34 | 25.24±4.05 | −10.402 | <0.001* |
| Smoking (%) | 48.4 | 40.7 | 8.431 | 0.004* |
| Drinking (%) | 32.3 | 30.3 | 0.651 | 0.454 |
| Diabetes (%) | 25.3 | 14.0 | 28.242 | <0.001* |
| College or above (%) | 55.1 | 47.0 | 9.287 | 0.003* |
| SBP (mmHg) | 130.98±18.62 | 119.73±15.75 | 18.637 | <0.001* |
| DBP (mmHg) | 79.527±12.28 | 74.56±10.62 | −8.100 | <0.001* |
| PR (bpm) | 78.77±11.92 | 78.44±12.21 | −0.505 | 0.614 |
| UA (mmol/L) | 324.54±92.65 | 307.52±83.70 | −3.608 | <0.001* |
| FBG (mmol/L) | 7.12±3.29 | 6.73±3.29 | −2.231 | 0.026* |
| TG (mmol/L) | 2.13±1.73 | 1.83±1.20 | −3.657 | <0.001* |
| TC (mmol/L) | 4.29±1.14 | 4.34±1.13 | 0.880 | 0.379 |
| HDL-C (mmol/L) | 1.01±0.30 | 1.06±0.30 | 2.862 | 0.004* |
| LDL-C (mmol/L) | 2.95±0.87 | 2.79±0.92 | −3.320 | 0.001* |
| ApoA1 (g/L) | 1.19±0.27 | 1.18±0.25 | −0.765 | 0.445 |
| ApoB (g/L) | 0.86±0.27 | 0.88±0.26 | 1.318 | 0.188 |
| Lp(a) (mg/L) | 217.49±202.60 | 214.03±202.21 | −0.319 | 0.750 |
Continuous parametric variables are reported as the mean ± SD. The Student t-test was used to compare quantitative traits that followed a normal distribution. Categoric variables were compared using chi-squared tests. *, statistically significant at P<0.05. EHT, essential hypertension; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; PR, pulse rate; UA, uric acid; FBG, fasting blood glucose; TG, triglyceride; TC, total cholesterol; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; Apo, apolipoprotein; Lp(a), lipoprotein A; SD, standard deviation
The genotype distributions of all polymorphisms in the cases and controls are shown in Table 2. The rs662799 SNP accorded with the HWE. The results indicated that the rs662799 polymorphism was significantly associated with EHT in codominant (OR =1.325, 95% CI: 1.062–1.654; P=0.013), dominant (OR =1.350, 95% CI: 1.091–1.671; P=0.006), and overdominant models (OR =0.781, 95% CI: 0.628–0.972; P=0.026). The frequency of the G allele of rs662799 was found to be significantly higher in the case group compared to the control group (OR =1.281, 95% CI: 1.075–1.528; P=0.006).
Table 2
| Genotype/allele | Case | Control | χ² | P value | OR (95% CI) |
|---|---|---|---|---|---|
| Codominant model | |||||
| AA | 383 | 434 | – | – | – |
| AG | 276 | 236 | 6.218 | 0.013* | 1.325 (1.062–1.654) |
| GG | 41 | 30 | 3.092 | 0.079 | 1.549 (0.948–2.529) |
| Dominant model | |||||
| AA | 383 | 434 | – | – | – |
| AG + GG | 317 | 266 | 7.645 | 0.006* | 1.350 (1.091–1.671) |
| Recessive model | |||||
| AA + AG | 659 | 670 | – | – | – |
| GG | 41 | 30 | 1.795 | 0.180 | 1.389 (0.857–2.252) |
| Over-dominant model | |||||
| AG | 276 | 236 | – | – | – |
| AA + GG | 424 | 464 | 4.927 | 0.026* | 0.781 (0.628–0.972) |
| Allele | |||||
| A | 1042 | 1,104 | – | – | – |
| G | 358 | 296 | 7.669 | 0.006* | 1.281 (1.075–1.528) |
*, statistically significant at P<0.05. APOA5, apolipoprotein A5; EHT, essential hypertension; OR, odds ratio; CI, confidence interval.
Furthermore, a multivariate logistic regression model was employed to assess the association between APOA5 rs662799 and the risk of EHT (Table 3). The analysis revealed that the G allele of rs662799 was an independent risk factor for EHT. After adjustments were made for confounding factors, including age, BMI, smoking, diabetes, UA, FBG, plasma TG, HDL-C, and LDL-C, and education level, the difference remained significant (OR =1.519, 95% CI: 1.203–1.917; P<0.001).
Table 3
| Variables | β | SE | Wald | OR | 95% CI | P value |
|---|---|---|---|---|---|---|
| Age (years) | 0.058 | 0.007 | 67.596 | 1.060 | 1.045–1.075 | <0.001* |
| BMI (kg/m2) | 0.150 | 0.017 | 80.505 | 1.162 | 1.124–1.200 | <0.001* |
| Smoking (%) | 0.475 | 0.130 | 13.338 | 1.608 | 1.246–2.075 | <0.001* |
| Diabetes (%) | 0.680 | 0.182 | 13.932 | 1.973 | 1.381–2.819 | <0.001* |
| College or above (%) | 0.379 | 0.123 | 9.553 | 1.461 | 1.149–1.858 | 0.002* |
| UA (mmol/L) | 0.001 | 0.001 | 2.893 | 1.001 | 1.000–1.003 | 0.089 |
| FBG (mmol/L) | −0.035 | 0.022 | 2.610 | 0.966 | 0.926–1.007 | 0.106 |
| TG (mmol/L) | 0.093 | 0.042 | 4.767 | 1.097 | 1.010–1.192 | 0.029* |
| HDL-C (mmol/L) | −0.241 | 0.216 | 1.238 | 0.786 | 0.514–1.201 | 0.266 |
| LDL-C (mmol/L) | 0.209 | 0.068 | 9.464 | 1.232 | 1.079–1.407 | 0.002* |
| G allele | 0.418 | 0.119 | 12.331 | 1.519 | 1.203–1.917 | <0.001* |
*, statistically significant at P<0.05. SE, standard error; OR, odds ratio; CI, confidence interval; BMI, body mass index; UA, uric acid; FBG, fasting blood glucose; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol.
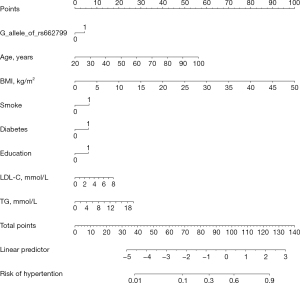
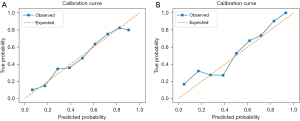
The results from multivariate logistic regression analysis showed that the APOA5 rs662799 G allele, age, BMI, smoking, diabetes, education, LDL-C, and TG were independent risk factors. Based on this result, we developed a risk evaluation model named GABSDTEL. The risk score can be calculated with the formula: GABSDTEL score = −8.67 + 0.06 × age + 0.16 × BMI + 0.50 × smoking + 0.47 × diabetes + 0.47 × education level + 0.11 × TG + 0.17 × LDL-C + 0.34 × G allele. The predictive nomogram of EHT in Figure 2 was constructed by combining the above independent prediction variables. As shown in Figure 3, the model demonstrated strong predictive performance, achieving AUC values of 0.722 (95% CI: 0.693–0.752; P<0.001) and 0.747 (95% CI: 0.690–0.804; P<0.001) in the training and validation sets, respectively. The Hosmer-Lemeshow tests (Figure 4) for calibration analysis showed no statistical significance in both of the training set (P=0.969) and the validation set (P=0.761), suggesting that the model has an excellent calibration ability.
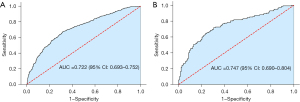
Based on the ROC curve analysis, the Youden index was calculated to be 0.413, with a sensitivity of 0.718 and a specificity of 0.695. The optimal threshold value was 0.486, which coincided with a total point value of approximately 96 on the nomogram. Notably, the nomogram exhibited the best predictive performance when setting the threshold at this point for dividing high-risk and low-risk groups.
Discussion
Our study showed that the G allele of APOA5 rs662799 was significantly associated with EHT in a Chinese population. Moreover, a highly discriminative and well-calibrated diagnostic model consisting of the selected clinical variables and the G allele of rs662799 was developed to predict the risk of EHT. To the best of our knowledge, this study is the first of its kind to develop such a model for EHT in a Chinese population.
The APOA5 gene is one of the most distinctive members of the APOA1/C3/A4/A5 gene cluster. It has been identified as a vital regulatory factor in TG metabolism and plasma lipid levels (15-17). The mechanisms by which APOA5 influences TG metabolism involve different processes. The APOA5 gene encodes an apolipoprotein that is involved in regulating the activity of lipoprotein lipase (LPL), which in turn reduces the synthesis of VLDL and promotes its hydrolysis (8). Among the common APOA5 SNPs reported in different populations, rs662799 is considered to be a major tagging SNP. The rs662799 polymorphism has been extensively investigated due to its correlation with elevated levels of TG, TC, and LDL-C and with decreased levels of HDL-C (18-20).
EHT is a multifaceted condition that is influenced by the synergistic interplay of genetic and environmental factors. As an important component of metabolic syndrome, the occurrence and progression of EHT are closely related to obesity, dyslipidemia, and diabetes, among other conditions. A recent study showed that the rs662799 polymorphism of the APOA5 gene is related to the incidence of myocardial infarction (13). Considering the close relations of this variant of the APOA5 gene with cardiovascular events and metabolic syndrome, we hypothesized that the rs662799 polymorphism and EHT were associated.
In a prospective cohort study lasting 8 years (21), Yamada et al. examined the correlation of five candidate gene polymorphisms with blood pressure and EHT incidence in a population of 2,267 Japanese participants. The findings revealed a significant correlation between the APOA5 rs662799 polymorphism and SBP and EHT incidence in men. Ouatou et al. conducted a study in a Moroccan population to examine the association between the APOA5 polymorphism and arterial hypertension (AHT). The results revealed a strong correlation between the −1131T>C (rs662799) polymorphism and AHT, indicating that this polymorphism is strongly correlated with increased SBP (22).
In our retrospective case-control study, we assessed the association of genetic variation in APOA5 rs662799 on the risk of EHT in a Chinese population. The results showed that the frequency of the G allele in rs662799 was significantly higher in the EHT group than in the control group. Multivariable analysis revealed a significant association between rs662799 and an increased susceptibility to EHT. Our findings thus suggest that the genetic polymorphism of the rs662799 variant in APOA5 may increase the risk of EHT by affecting lipid metabolism. Fatma et al. (23) discovered that the G allele of rs662799 reduces ribosomal translation efficiency, resulting in decreased APOA5 concentration, a subsequent lowering of LPL activity, an increase in VLDL production, and the inhibition of hydrolysis. Experimental evidence suggests that persistent hypertriglyceridemia causes endothelial dysfunction, characterized by reduced responsiveness to vasodilator stimuli and decreased nitric oxide availability, potentially leading to elevated blood pressure (22-24). In addition to affecting plasma lipid levels, the APOA5 gene is also linked to an elevated risk of metabolic syndrome (25,26), atherosclerosis (27), stroke (28), and coronary artery disease (29,30), all of which can further lead to the occurrence of EHT (31-33). Yin et al. discovered a noteworthy link between the APOA5 rs662799 polymorphism and the risk of T2DM. This suggests that the G allele independently contributes to the progression of T2DM (34). Chauhan et al. found that APOA5 rs662799 affects the incidence of obesity and further discovered that compared to noncarriers, carriers of the minor allele homozygote (rs662799, A-1131G) have a significantly higher BMI and serum TG levels but significantly lower serum HDLC levels (13). Moreover, reduced plasma APOA5 levels were observed in obese individuals from various countries and were negatively correlated with BMI (35). In a case-control study involving 631 Han Chinese individuals, Chen et al. demonstrated that only the APOA5 rs662799 variant was strongly associated with coronary artery disease among seven SNPs related to lipid metabolism pathways relevant to this disease (29).
Several limitations to this study should be mentioned. First, the study was limited by a small sample size and a single-center design; thus, the generalizability (external validity) of the study remains unclear. Second, the gender and age of the two groups were not fully matched. Third, this study did not conduct any biological functional verification. Therefore, further large-scale studies with adequate functional analysis are required to validate the findings of this study.
Conclusions
The rs662799 variant of APOA5 was significantly associated with susceptibility to EHT. We evaluated the correlation between rs662799 and EHT with a multivariate logistic regression model, and the results indicate that the G allele of rs662799 was an independent risk factor for EHT. The established nomogram and GABSDTEL score in our study may serve as a new risk predicting method for EHT in the Chinese population.
Acknowledgments
We would like to thank AME Editing Service for their help in polishing our paper.
Funding: This research was funded by Tianshan Elite Science and Technology Innovation Leading Talents Program of Xinjiang Uyghur Autonomous Region (High-level Leading Talents; grant No. 2022TSYCLJ0023).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-289/rc
Data Sharing Statement: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-289/dss
Peer Review File: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-289/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-289/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and received ethical approval from the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University (No. K202309-08). Informed consent was obtained from all research participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mills KT, Stefanescu A, He J. The global epidemiology of hypertension. Nat Rev Nephrol 2020;16:223-37. [Crossref] [PubMed]
- Mancia G, Kreutz R, Brunström M, et al. 2023 ESH Guidelines for the management of arterial hypertension The Task Force for the management of arterial hypertension of the European Society of Hypertension: Endorsed by the International Society of Hypertension (ISH) and the European Renal Association (ERA). J Hypertens 2023;41:1874-2071. [Crossref] [PubMed]
- Mills KT, Bundy JD, Kelly TN, et al. Global Disparities of Hypertension Prevalence and Control: A Systematic Analysis of Population-Based Studies From 90 Countries. Circulation 2016;134:441-50. [Crossref] [PubMed]
- Bigazzi R, Zagato L, Lanzani C, et al. Hypertension in High School Students: Genetic and Environmental Factors: The HYGEF Study. Hypertension 2020;75:71-8. [Crossref] [PubMed]
- Padmanabhan S, Dominiczak AF. Genomics of hypertension: the road to precision medicine. Nat Rev Cardiol 2021;18:235-50. [Crossref] [PubMed]
- de Luis Roman D, Primo D, Izaola O, et al. Association of the APOA-5 Genetic Variant rs662799 with Metabolic Changes after an Intervention for 9 Months with a Low-Calorie Diet with a Mediterranean Profile. Nutrients 2022;14:2427. [Crossref] [PubMed]
- Pennacchio LA, Olivier M, Hubacek JA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 2001;294:169-73. [Crossref] [PubMed]
- Wen Y, Chen YQ, Konrad RJ. The Regulation of Triacylglycerol Metabolism and Lipoprotein Lipase Activity. Adv Biol (Weinh) 2022;6:e2200093. [Crossref] [PubMed]
- Takanashi M, Kimura T, Li C, et al. Critical Role of SREBP-1c Large-VLDL Pathway in Environment-Induced Hypertriglyceridemia of Apo AV Deficiency. Arterioscler Thromb Vasc Biol 2019;39:373-86. [Crossref] [PubMed]
- Su X, Kong Y, Peng DQ. New insights into apolipoprotein A5 in controlling lipoprotein metabolism in obesity and the metabolic syndrome patients. Lipids Health Dis 2018;17:174. [Crossref] [PubMed]
- You Y, Wu YH, Zhang Y, et al. Effects of polymorphisms in APOA5 on the plasma levels of triglycerides and risk of coronary heart disease in Jilin, northeast China: a case-control study. BMJ Open 2018;8:e020016. [Crossref] [PubMed]
- Lin E, Kuo PH, Liu YL, et al. Detection of susceptibility loci on APOA5 and COLEC12 associated with metabolic syndrome using a genome-wide association study in a Taiwanese population. Oncotarget 2017;8:93349-59. [Crossref] [PubMed]
- Chauhan W, Fatma R, Wahab A, et al. Cataloging the potential SNPs (single nucleotide polymorphisms) associated with quantitative traits, viz. BMI (body mass index), IQ (intelligence quotient) and BP (blood pressure): an updated review. Egypt J Med Hum Genet 2022;23:57. [Crossref]
- Yang J, Wang X, Jiang S. Development and validation of a nomogram model for individualized prediction of hypertension risk in patients with type 2 diabetes mellitus. Sci Rep 2023;13:1298. [Crossref] [PubMed]
- Forte TM, Ryan RO. Apolipoprotein A5: Extracellular and Intracellular Roles in Triglyceride Metabolism. Curr Drug Targets 2015;16:1274-80. [Crossref] [PubMed]
- Gombojav B, Lee SJ, Kho M, et al. Multiple susceptibility loci at chromosome 11q23.3 are associated with plasma triglyceride in East Asians. J Lipid Res 2016;57:318-24. [Crossref] [PubMed]
- May-Zhang L, Liu M, Black D, et al. Apolipoprotein A5, a unique modulator of fasting and postprandial triglycerides. Biochim Biophys Acta Mol Cell Biol Lipids 2022;1867:159185. [Crossref] [PubMed]
- Wang Y, Lu Z, Zhang J, et al. The APOA5 rs662799 polymorphism is associated with dyslipidemia and the severity of coronary heart disease in Chinese women. Lipids Health Dis 2016;15:170. [Crossref] [PubMed]
- Halalkhor S, Jalali F, Tilaki KH, et al. Association of two common polymorphisms of apolipoprotein A5 gene with metabolic syndrome indicators in a North Iranian population, a cross-sectional study. J Diabetes Metab Disord 2014;13:48. [Crossref] [PubMed]
- Ovando Gómez V, Zavaleta Muñiz SA, Ochoa-Díaz-López H, et al. Association of rs662799 and rs5070 genetic polymorphisms with hypertriglyceridemia and atherogenic dyslipidemia in pediatric patients in Southeast Mexico. Clin Investig Arterioscler 2023;35:53-63. [Crossref] [PubMed]
- Yamada Y, Ando F, Shimokata H. Association of the genetic variants of APOA5 and PRKCH with hypertension in community-dwelling Japanese individuals. Mol Med Rep 2008;1:407-14. [Crossref] [PubMed]
- Ouatou S, Ajjemami M, Charoute H, et al. Association of APOA5 rs662799 and rs3135506 polymorphisms with arterial hypertension in Moroccan patients. Lipids Health Dis 2014;13:60. [Crossref] [PubMed]
- Fatma R, Chauhan W, Riyaz S, et al. Genetic association analysis of rs662799 (− 1131A > G) polymorphism of APOA5 gene with morphometric and physio-metric traits using multiplex PCR. Egypt J Med Hum Genet 2023;24:19. [Crossref]
- Kusterer K, Pohl T, Fortmeyer HP, et al. Chronic selective hypertriglyceridemia impairs endothelium-dependent vasodilatation in rats. Cardiovasc Res 1999;42:783-93. [Crossref] [PubMed]
- Masjoudi S, Sedaghati-Khayat B, Givi NJ, et al. Kernel machine SNP set analysis finds the association of BUD13, ZPR1, and APOA5 variants with metabolic syndrome in Tehran Cardio-metabolic Genetics Study. Sci Rep 2021;11:10305. [Crossref] [PubMed]
- Choi WJ, Shin D. Interactions between red and processed meat consumption and APOA5 gene variants associated with the incidence of metabolic syndrome in Korean adults. Genes Nutr 2022;17:5. [Crossref] [PubMed]
- Guardiola M, Cofán M, de Castro-Oros I, et al. APOA5 variants predispose hyperlipidemic patients to atherogenic dyslipidemia and subclinical atherosclerosis. Atherosclerosis 2015;240:98-104. [Crossref] [PubMed]
- Au A, Griffiths LR, Irene L, et al. The impact of APOA5, APOB, APOC3 and ABCA1 gene polymorphisms on ischemic stroke: Evidence from a meta-analysis. Atherosclerosis 2017;265:60-70. [Crossref] [PubMed]
- Chen H, Ding S, Zhou M, et al. Association of rs662799 in APOA5 with CAD in Chinese Han population. BMC Cardiovasc Disord 2018;18:2. [Crossref] [PubMed]
- Do R, Stitziel NO, Won HH, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015;518:102-6. [Crossref] [PubMed]
- Jayedi A, Rashidy-Pour A, Khorshidi M, et al. Body mass index, abdominal adiposity, weight gain and risk of developing hypertension: a systematic review and dose-response meta-analysis of more than 2.3 million participants. Obes Rev 2018;19:654-67. [Crossref] [PubMed]
- Leong XF, Ng CY, Jaarin K. Animal Models in Cardiovascular Research: Hypertension and Atherosclerosis. Biomed Res Int 2015;2015:528757. [Crossref] [PubMed]
- Malakar AK, Choudhury D, Halder B, et al. A review on coronary artery disease, its risk factors, and therapeutics. J Cell Physiol 2019;234:16812-23. [Crossref] [PubMed]
- Yin YW, Sun QQ, Wang PJ, et al. Genetic polymorphism of apolipoprotein A5 gene and susceptibility to type 2 diabetes mellitus: a meta-analysis of 15,137 subjects. PLoS One 2014;9:e89167. [Crossref] [PubMed]
- Su X, Peng D. The exchangeable apolipoproteins in lipid metabolism and obesity. Clin Chim Acta 2020;503:128-35. [Crossref] [PubMed]


