
Multinational experience with next-generation sequencing: opportunity to identify transthyretin cardiac amyloidosis and Fabry disease
Highlight box
Key findings
• A 17-gene next-generation sequencing (NGS) panel identified hypertrophic cardiomyopathy (HCM) variants (21.5% of 535 samples) and HCM phenocopy variants (3.2%) among patients with unexplained increased left ventricular wall thickness (LVWT), with an overall diagnostic yield of 24.7%.
What is known and what is new?
• A genetic diagnosis for patients with LVWT of unknown origin by either diagnostic genetic testing or predictive genetic testing aids in determining the prognosis and allows for timely management.
• The 17-gene NGS panel, using dried blood spot samples, effectively detected gene variants that cause the most common forms of HCM and HCM phenocopies, including the treatable disorders transthyretin cardiac amyloidosis and Fabry disease, in patients with increased LVWT from six countries.
What is the implication, and what should change now?
• This 17-gene NGS screening strategy has the potential to reduce the diagnostic challenges experienced by patients with unexplained increased LVWT and facilitate personalized care.
Introduction
The 2020 American College of Cardiology/American Heart Association guidelines define hypertrophic cardiomyopathy (HCM) as “a maximal end-diastolic wall thickness of ≥15 mm anywhere in the left ventricle in the absence of another cause of hypertrophy in adults” (1). The hallmark of HCM is its clinical heterogeneity, ranging from a benign asymptomatic state to arrhythmias, heart dysfunction, and sudden cardiac death (SCD) (2). In young adults, the most common etiology of HCM is SCD (3). The global prevalence of HCM among adults is 0.16–0.29% (~1:625–1:344 individuals). An even higher prevalence of 0.6% (1:167) has been reported with sensitive radiological imaging techniques, evaluation of family members, and greater use of genetic testing (4).
Variations in genes encoding cardiac sarcomere proteins account for up to 60% of HCM cases in adults (5). HCM is regarded as a monogenic cardiac disease; however, it exhibits extensive genetic heterogeneity. Due to this genetic diversity, the pathogenic genes remain unidentified in approximately 40% of HCM patients (2,5). Many genes other than sarcomeric genes have been associated with HCM; therefore, HCM is no longer considered monogenic in origin and may be due to polygenicity or oligogenicity (6,7).
Certain diseases that mimic HCM are referred to as HCM phenocopies (2). These include conditions such as protein kinase adenosine monophosphate-activated noncatalytic subunit gamma 2 (PRKAG2) cardiomyopathy, glycogen storage disorders such as Danon disease, cardiac amyloidosis, lysosomal storage disorders [e.g. Fabry disease (FD)], and other inborn errors of metabolism (4). Transthyretin cardiac amyloidosis (ATTR-CA) and FD are caused by pathogenic variants of TTR (18q12.1) and GLA (Xq22.1), respectively (8-10), and increased left ventricular wall thickness (LVWT) is a key diagnostic feature of both disorders (8-11). Amyloid cardiomyopathy results from extracellular deposits of amyloid in the myocardium, leading to symptoms of chest pain, arrhythmia, and sudden death (10). Clinically, FD may present as cardiac hypertrophy, arrhythmias, fibrosis, and heart failure due to reduced α-galactosidase A enzyme activity (8). It is crucial to differentiate sarcomeric HCM and phenotypically similar conditions because the prognosis and management differ greatly as cause-specific therapies are available (12,13). Hence, clinicians should consider the presence of HCM phenocopies in patients with unexplained left ventricular hypertrophy (LVH) who do not meet the standard HCM diagnostic criteria, as well as predictive genetic testing in relatives of a patient with confirmed HCM or more limited hypertrophy (13–14 mm) (1). Our study explored the feasibility and practicality of the application of a 17-gene next-generation sequencing (NGS) panel to detect gene variants that cause the most common forms of HCM and HCM phenocopies in patients with unexplained increased LVWT from six countries and detection rates for pathogenic/likely pathogenic variants are reported.
Methods
Sample population
The study population for this cross-sectional study included subjects from cardiology clinics located in Colombia, Brazil, Mexico, Turkey, Israel, and Saudi Arabia. Countries were selected based on interest in participation, the potential pool of undiagnosed patients in the cardiology domain, and the ability to execute the study. All participating institutions were informed and agreed to the study. From the study population, 544 samples were received by the Diagnósticos Laboratoriais Especializados (DLE) between May 2019 and October 2020. However, due to preanalytical issues and the absence of informed consent, only 535 samples were analyzed for this study.
Inclusion criteria
Patients with an increased LVWT of ≥13 mm in one or more left ventricular myocardial segments (as measured by any imaging method—computed tomography, cardiac magnetic resonance imaging, or echocardiography), in the absence of abnormal loading disease conditions that justify increased LVWT, were included in the study.
Exclusion criteria
Patients with increased LVWT due to severe hypertension (defined as systolic blood pressure ≥180 mmHg and/or diastolic blood pressure ≥120 mmHg), severe aortic stenosis (≤1 cm2), or genetically confirmed etiology of increased LVWT were excluded from the study.
Ethical approval and patient consent to participate
This study was performed in accordance with the ethical principles of the Declaration of Helsinki (as revised in 2013). The central laboratory study was approved by Institute of Childcare and Pediatrics Martagão Gesteira/Federal University of Rio de Janeiro under approval No. CAAE 53630421.3.0000.5264. Signed informed consent forms for blood sampling and analysis were obtained at the participating sites from all patients included in the study.
Procedures
Peripheral dried blood spot (DBS) samples were collected from patients onto a filter paper and stored at room temperature. Demographic details, including age, gender, and country of origin, were simultaneously recorded. Clinical and cardiac imaging data of patients were not collected in this pilot study. Deoxyribonucleic acid (DNA) from blood spots was extracted automatically using a QIAsymphony Investigator Kit® (Qiagen®). Both DBS and extracted DNA were kept refrigerated (4 ℃). Anonymized samples were analyzed at the DLE.
Sequencing analysis
In selecting genes to be included in the NGS panel, the global prevalence, national and regional epidemiology, and local technical capabilities were considered. The pathogenicity of variants was classified according to the consensus recommendations published by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) (14,15) into categories such as pathogenic, likely pathogenic (probability greater than 90% of being pathogenic), uncertain significance, likely benign (more than 90% likely to be benign), and benign. ACMG uses a series of criteria to establish a scoring system based on variant information (e.g., protein effect, information in literature, position in the transcript, functional assays, prediction software, and database). The variant classification is determined based on the presence or absence of certain traits. A 17-gene panel was developed to detect known splice regions and flanking regions (20 base pairs adjacent to each exon) of the targeted genes using DBS samples. Information on the 17 genes included in the panel is presented in Table 1.
Table 1
| Gene | Locus | Exons sequenced | OMIM | HCM or HCM phenocopy |
|---|---|---|---|---|
| ACTC1 | 15q14 | 7 | 102540 | HCM |
| TNNI3 | 19q13.42 | 8 | 191044 | HCM |
| MYBPC3 | 11p11.2 | 35 | 600958 | HCM |
| MYH7 | 14q11.2 | 40 | 160760 | HCM |
| MYL2 | 12q24.11 | 7 | 160781 | HCM |
| MYL3 | 3p21.31 | 7 | 160790 | HCM |
| TPM1 | 15q22.2 | 14 | 191010 | HCM |
| TNNT2 | 1q32.1 | 17 | 191045 | HCM |
| TNNC1 | 3p21.1 | 6 | 191040 | HCM |
| PRKAG2 | 7q36.1 | 22 | 602743 | HCM phenocopy |
| PTPN11 | 12q24.13 | 16 | 176876 | HCM phenocopy |
| TTR | 18q12.1 | 4 | 176300 | HCM phenocopy |
| LAMP2 | Xq24 | 10 | 309060 | HCM phenocopy |
| GLA | Xq22.1 | 7 | 300644 | HCM phenocopy |
| PLN | 6q22.31 | 2 | 172405 | HCM phenocopy |
| FLNC | 7q32.1 | 48 | 102565 | HCM phenocopy |
| DES | 2q35 | 9 | 125660 | HCM phenocopy |
OMIM, Online Mendelian Inheritance in Man; HCM, hypertrophic cardiomyopathy.
Library preparation and NGS
Initially, the dilution of genomic DNA (10–500 ng) was performed with ultrapure water. Genomic libraries were prepared according to instructions provided by Agilent (Agilent Sure Select XT HS and XT Low Input Custom 1-499 kb 96 reactions Design ID: 3223981). Qualitative and quantitative validation of all DNA libraries was performed using an automatic gel electrophoresis method (TapeStation D1000 Screen Tape) and a fluorescence-based measurement method (Qubit™ dsDNA HS Assay Kit), respectively.
SureSelect XT HS and XT Low Input were used for pooling and hybridizing successfully validated DNA libraries to synthesize probes that were in accordance with the human genome reference sequence version GRCh37 (hg19). In-house-designed, biotin-labeled oligonucleotide probes were generated for binding with targeted gene sequences.
The Agilent SureSelect XT HS and XT Low Input preparation automation guide (Agilent) was utilized to prepare the target-captured library as per the manufacturer’s protocol, with slight modifications for samples on DBS. In addition, the automatic gel electrophoresis method (TapeStation D1000 Screen Tape) was employed to identify the final, size-adjusted concentration of the target-captured DNA fragments.
The sequencing of target-enriched DNA fragments was performed by Illumina NextSeq 500 and NovaSeq 6000. These sequencers carried out the sequencing via pair-end and two 150-base pair reads having an average coverage of a minimum of 20× to ensure the quality of the data. A fully characterized positive control was included in each sequencing run.
NGS quality and data analysis
High-end quality control (QC) procedures were conducted along the sequencing process to identify the samples. Apart from sample identification, these procedures were also performed to ensure good quality of isolated DNA, library preparation, target capture, and sequencing. Furthermore, mapping and variant QC metrics were computed and used on the sequencing output to exclude and rerun failed samples. A bioinformatics analysis pipeline was utilized to align the sequencing data against the human genome reference sequence version GRCh37 (hg19) and obtain relevant information. An in-house software (‘DLE-Tool’) that uses Burrows-Wheeler for alignment (0.7.15), variant calling (single-nucleotide variants and indels) based on mpileup (1.3.1), and Ensembl Variant Effect Predictor (v105) annotation was employed to assess the pathogenicity [or variant of uncertain significance (VUS)] of all identified variants. Franklin variant interpretation (Genoox) was used especially in cases with novel variants.
Statistical analysis
Demographic and gene variant data were summarized using descriptive statistics.
Continuous variables were described using mean with SD and median with range, and N was calculated for categorical variables.
Results
A total of 535 samples from patients across Colombia, Brazil, Mexico, Turkey, Israel, and Saudi Arabia were analyzed [311 (58.1%) males, 222 (41.5%) females, and gender not recorded for 2 (0.4%) individuals]. Table 2 provides the demographic characteristics of the included patients.
Table 2
| Overalla | HCM + HCM phenocopies | HCMb | HCM phenocopiesc | ATTR-CA | FD | |
|---|---|---|---|---|---|---|
| N | 535 | 128 | 113 | 17 | 7 | 2 |
| Mean age (SD), years | 49.9 (17.7) | 42.8 (17.9) | 42.0 (17.4) | 46.5 (20.1) | 54.6 (17.0) | 69.0 (1.4) |
| Median age [range], years | 51 [4–92] | 43 [4–85] | 43 [4–85] | 46 [11–73] | 44 [31–73] | 69 [68–70] |
a, excluded were samples with missing (n=5) or incorrect age (n=5); b, HCM variants: MYBPC3, MYH7, TNNI3, TNNT2, TNNC1, TPM1, MYL3, MYL2, and ACTC1; c, HCM phenocopy variants (this includes two patients with double mutations, each with an HCM variant and an HCM phenocopy): DES, GLA, FLNC, LAMP2, PLN, PRKAG2, PTPN11, and TTR. HCM, hypertrophic cardiomyopathy; ATTR-CA, transthyretin cardiac amyloidosis; FD, Fabry disease; SD, standard deviation.
HCM and HCM phenocopy variants considered to be pathogenic/likely pathogenic were identified in a total of 128 patient samples (23.9%). In four of the 128 samples (0.7%), a second pathogenic/likely pathogenic variant (associated with HCM in two samples and with an HCM phenocopy in two samples) was detected in addition to the HCM variant. Among the 132 detected variants, 115 (21.5%) variants were associated with HCM and 17 (3.2%) variants with HCM phenocopies. The mean [standard deviation (SD)] age of the 128 patients was 42.8 (17.9) years. The distribution of variants by country is shown in Figure 1, and classifications of pathogenicity and nucleotide and amino acid changes are provided in Table S1.
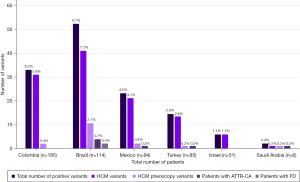
Of the patients with pathogenic/likely pathogenic HCM variants (n=113), 56 patients (49.6%) were male, and 57 patients were female (50.4%). Overall, the most prevalent HCM variants were MYH7 (n=60, 11.2%) and MYBPC3 (n=41, 7.7%) (Figure 2).
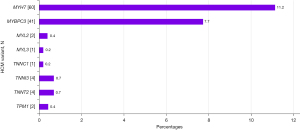
HCM phenocopy variants considered to be pathogenic/likely pathogenic were identified in 17 patients [9 males (52.9%) and 8 females (47.1%)]. TTR variants were the most prevalent HCM phenocopy variants (n=7; 1.3%) and were found in 5 males and 2 females [mean (SD) age 54.6 (17.0) years]. Other pathogenic/likely pathogenic HCM phenocopy variants detected in more than one sample included LAMP2 (n=3, 0.6%), DES (n=2, 0.4%), and GLA variants (n=2, 0.4%). The GLA variants were found in female patients [mean (SD) age 69.0 (1.4) years] and were predicted to be associated with the ‘classic’ phenotype (c.413delG) and a ‘later-onset’ phenotype of FD (c.644A>G) according to the International Fabry Disease Genotype–Phenotype (dbFGP) database (http://dbfgp.org/dbFgp/fabry/Mutation.html), which was available online during the assessment of data.
Discussion
HCM is the most common inherited form of cardiomyopathy and is associated with considerable genotypic variations. Advances in gene sequencing provide opportunities to improve diagnostic certainty and differentiate genetic sarcomeric HCM from phenocopies (16). However, as a result of an incomplete understanding of the genetic variants and their pathogenicity, patients may remain with a diagnosis unconfirmed by molecular diagnostics (5). Establishing an early diagnosis is essential as it allows the determination of the prognosis and appropriate therapeutic intervention. In this pilot study of 535 patients with unexplained increased LVWT, the 17-gene NGS panel showed a diagnostic yield of 24.7% for pathogenic/likely pathogenic variants associated with HCM or HCM phenocopies.
The diagnostic yield of the panel for confirmed HCM was found to be 21.5%. Evidence from published studies presented in Table S2 indicates that the diagnostic yield of NGS screening methods for HCM ranged from 21% to 79% (17-26). The most prevalent HCM gene variants in the present study were MYH7 (11.2%) and MYBPC3 (7.7%). HCM phenocopies were confirmed in 3.2% of patients, which is less frequent compared with the published range of 5–10% for testing projects for HCM phenocopies (27). The present study also reported ATTR-CA as the most frequent phenocopy (1.3%). Six of the published studies investigated the diagnostic yield for ATTR-CA among patients with HCM (Table S2). Five studies did not detect variants in the TTR gene (17,18,20-22). A study conducted in Italy included 343 patients who had been referred with a tentative HCM diagnosis and reported a diagnostic yield of 3.5% and 2% for ATTR-CA and FD, respectively (11-gene panel) (26). The diagnostic yield for FD (0.4% in the current study) ranged from 0% to 5.6% in NGS-based studies among HCM patients (Table S2) (17-20,22,24-26). Less stringent risk stratification and greater geographic dispersion of patients in the current study may have contributed to the differences in the diagnostic yield rates compared with the other studies discussed above.
It is noteworthy that the present study enrolled the highest number of patients compared to other published NGS-based studies that analyzed HCM patients as summarized in Table S2. Additionally, it is also the only study that enrolled patients from different countries. Interestingly, the gene panels used in three of the ten studies did not include the TTR gene. The results of the present study signify the importance of the inclusion of TTR in the NGS screening panels.
Hereditary ATTR-CA is an autosomal-dominant disease with considerable phenotypic heterogeneity with certain TTR variants causing exclusively infiltrative cardiomyopathy (10). FD is an X-linked disorder with phenotypes varying from the classic phenotype, with pediatric-onset and multi-organ involvement, to later-onset, a predominantly cardiac phenotype. Manifestations are diverse in female FD patients in part due to variations in residual enzyme activity and X chromosome inactivation patterns (13). Lack of recognition of these clinical entities, non-specific symptoms, and co-morbidities often lead to delayed diagnosis, resulting in disease progression (10,13).
A confirmed diagnosis of ATTR-CA or FD allows appropriate and timely initiation of cause-specific therapy (i.e. stabilizing molecules or genetic silencers for ATTR-CA (12), and enzyme replacement therapy or chaperone therapy for FD (13,28) to relieve symptoms and to improve the prognosis.
The diagnostic yield for pathogenic variants and VUS is enhanced by using the NGS-based technique. The clinical interpretation of NGS testing results was based on the pathogenicity of the sequencing variants, which was ascertained as per the ACMG/AMP guidelines (14,15). Additionally, a VUS temperature scale may be used to assess the variant classification (14). Other web-based databases of genetic variants that may contain additional disease-relevant information on VUS are presented in Table S3. These resources serve to support clinicians in the interpretation of genetic testing results, aiding in the accurate assessment and understanding of genetic variants associated with HCM and HCM phenocopies.
This pilot study provided the first evidence of the feasibility of NGS testing using the 17-gene panel and its effectiveness in increasing the diagnostic yield for HCM and its phenocopies, including ATTR-CA and FD. Therefore, it has the potential to reduce the diagnostic odyssey experienced by patients with an unexplained increased LVWT and their families and facilitate appropriate genetic counseling (including identification of relatives at risk of having an inherited heart condition) and a better outcome of current therapies (29).
An important limitation of this pilot study is that the clinical data of patients were not collected. Furthermore, cardiac imaging data were not collected because of a lack of access to cardiac magnetic resonance imaging in the participating countries. Thus, we were not able to perform critical analyses of genotype–phenotype associations. Data on the ethnicity of patients were not collected. Such data are important as variations in ethnic background affect the penetrance and expressivity of genetic variants. The gene panel used in our study was limited to 17 genes associated with the most common forms of HCM and HCM phenocopies. This approach challenged the detection of other genes, including novel genes and deep intronic variants. Lastly, the number of patients included in the current pilot study was relatively small. A larger confirmatory study involving 2,068 patients from 21 countries is currently ongoing.
Conclusions
A diagnostic pilot study was conducted to investigate the feasibility of using a 17-gene NGS panel to detect gene variants associated with the most common forms of HCM and HCM phenocopies in patients with an unexplained increased LVWT. DBS samples derived from 535 patients from six countries were analyzed. This strategy was effective in the identification of patients with pathogenic/likely pathogenic variants associated with HCM (21.5%) or HCM phenocopies (3.2%) with an overall diagnostic yield of 24.7%. Early confirmation of the underlying etiology of HCM and HCM phenocopies is essential as patient outcomes largely depend on the early initiation of cause-specific therapy, if available.
Acknowledgments
We thank the patients, clinicians, and hospital staff who were involved in this NGS diagnostic project, which was funded by Sanofi. BioQuest Solutions provided editorial support for the development of this manuscript, which was funded by Sanofi.
Funding: This diagnostic project was funded by
Footnote
Data Sharing Statement: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-191/dss
Peer Review File: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-191/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-23-191/coif). All authors report this study was funded by Sanofi. S.M.e.S. received speaker honoraria and grant/research support from Takeda, Pint Pharma, Pfizer and Sanofi; consulting fees from Chiesi Farmaceutici; and support for attending meetings and traveling from Takeda, Pint Pharma, Pfizer and Sanofi; and received support from Sanofi for current research and manuscript. A.V.F.C. received advisory fees and honoraria for lectures from Takeda, Sanofi and Pint-Pharma; support for attending meetings and/or traveling from Sanofi, Takeda and Pint-Pharma; and grant/research support from Multicare; Participation on a Data Safety Monitoring Board or Advisory Board for Takeda, Sanofi, and Pint-Pharma; and received support from Sanofi for current research and manuscript. M.A. received speaker honoraria from Takeda, Pfizer, and Sanofi; and grant/research support from Novartis and Pfizer; and received support from Sanofi for current research and manuscript. J.P.C. received support from Sanofi for current research and manuscript. A.A.d.F. received support from Sanofi for current research and manuscript. A.F. is an employee and shareholder of Sanofi and received support from Sanofi for current research and manuscript. J.E.G.M. received support from Sanofi for current research and manuscript. F.J.M.G. received support from Sanofi for current research and manuscript. O.C. received advisory fees from Sanofi Aventis and received support from Sanofi for current research and manuscript. I.M. is an employee and shareholder of Sanofi and received support from Sanofi for current research and manuscript. M.M. is an employee and shareholder of Sanofi and received support from Sanofi for current research and manuscript. C.M. received support from Sanofi for current research and manuscript. J.L.B.P. received advisory fees from Sanofi and received support from Sanofi for current research and manuscript. M.G.R. received support from Sanofi for current research and manuscript. M.J.R.G. received advisory fees and honoraria for lectures from Sanofi and received support from Sanofi for current research and manuscript. O.T. receives honoraria and lecture fees from Pfizer and Sanofi, payment for expert testimony from Pfizer; and received support from Sanofi for current research and manuscript. H.O. received speaker honoraria from Sanofi and received support from Sanofi for current research and manuscript. The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ommen SR, Mital S, Burke MA, et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2020;142:e558-631. [PubMed]
- Bagnall RD, Ingles J, Dinger ME, et al. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients With Hypertrophic Cardiomyopathy. J Am Coll Cardiol 2018;72:419-29. [Crossref] [PubMed]
- Maron BJ, Rowin EJ, Maron MS. Paradigm of Sudden Death Prevention in Hypertrophic Cardiomyopathy. Circ Res 2019;125:370-8. [Crossref] [PubMed]
- Marian AJ, Braunwald E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ Res 2017;121:749-70. [Crossref] [PubMed]
- Teekakirikul P, Zhu W, Huang HC, et al. Hypertrophic Cardiomyopathy: An Overview of Genetics and Management. Biomolecules 2019;9:878. [Crossref] [PubMed]
- Baulina NM, Kiselev IS, Chumakova OS, et al. Hypertrophic cardiomyopathy as an oligogenic disease: transcriptomic arguments. Mol Biol (Mosk) 2020;54:955-67. [PubMed]
- Coppini R, Santini L, Olivotto I, et al. Abnormalities in sodium current and calcium homoeostasis as drivers of arrhythmogenesis in hypertrophic cardiomyopathy. Cardiovasc Res 2020;116:1585-99. [Crossref] [PubMed]
- Linhart A, Germain DP, Olivotto I, et al. An expert consensus document on the management of cardiovascular manifestations of Fabry disease. Eur J Heart Fail 2020;22:1076-96. [Crossref] [PubMed]
- Pieroni M, Moon JC, Arbustini E, et al. Cardiac Involvement in Fabry Disease: JACC Review Topic of the Week. J Am Coll Cardiol 2021;77:922-36. [Crossref] [PubMed]
- Yamamoto H, Yokochi T. Transthyretin cardiac amyloidosis: an update on diagnosis and treatment. ESC Heart Fail 2019;6:1128-39. [Crossref] [PubMed]
- Beneyto M, Cariou E, Brunel J, et al. Tip of the iceberg: a tertiary care centre retrospective study of left ventricular hypertrophy aetiologies. Open Heart 2021;8:e001462. [Crossref] [PubMed]
- Garcia-Pavia P, Rapezzi C, Adler Y, et al. Diagnosis and treatment of cardiac amyloidosis: a position statement of the ESC Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2021;42:1554-68. [Crossref] [PubMed]
- Ortiz A, Germain DP, Desnick RJ, et al. Fabry disease revisited: Management and treatment recommendations for adult patients. Mol Genet Metab 2018;123:416-27. [Crossref] [PubMed]
- Ellard S, Baple EL, Callaway A, et al. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020. Available online: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf. Accessed on: 20 February 2023.
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Bonaventura J, Polakova E, Vejtasova V, et al. Genetic Testing in Patients with Hypertrophic Cardiomyopathy. Int J Mol Sci 2021;22:10401. [Crossref] [PubMed]
- Jääskeläinen P, Vangipurapu J, Raivo J, et al. Genetic basis and outcome in a nationwide study of Finnish patients with hypertrophic cardiomyopathy. ESC Heart Fail 2019;6:436-45. [Crossref] [PubMed]
- Tran Vu MT, Nguyen TV, Huynh NV, et al. Presence of Hypertrophic Cardiomyopathy Related Gene Mutations and Clinical Manifestations in Vietnamese Patients With Hypertrophic Cardiomyopathy. Circ J 2019;83:1908-16. [Crossref] [PubMed]
- Zhao Y, Feng Y, Ding X, et al. Identification of a novel hypertrophic cardiomyopathy-associated mutation using targeted next-generation sequencing. Int J Mol Med 2017;40:121-9. [Crossref] [PubMed]
- Hayashi T, Tanimoto K, Hirayama-Yamada K, et al. Genetic background of Japanese patients with pediatric hypertrophic and restrictive cardiomyopathy. J Hum Genet 2018;63:989-96. [Crossref] [PubMed]
- Bonaventura J, Norambuena P, Tomašov P, et al. The utility of the Mayo Score for predicting the yield of genetic testing in patients with hypertrophic cardiomyopathy. Arch Med Sci 2019;15:641-9. [Crossref] [PubMed]
- Nagyova E, Radvanszky J, Hyblova M, et al. Targeted next-generation sequencing in Slovak cardiomyopathy patients. Bratisl Lek Listy 2019;120:46-51. [Crossref] [PubMed]
- Norrish G, Jager J, Field E, et al. Yield of Clinical Screening for Hypertrophic Cardiomyopathy in Child First-Degree Relatives. Circulation 2019;140:184-92. [Crossref] [PubMed]
- Rubattu S, Bozzao C, Pennacchini E, et al. A Next-Generation Sequencing Approach to Identify Gene Mutations in Early- and Late-Onset Hypertrophic Cardiomyopathy Patients of an Italian Cohort. Int J Mol Sci 2016;17:1239. [Crossref] [PubMed]
- Cecconi M, Parodi MI, Formisano F, et al. Targeted next-generation sequencing helps to decipher the genetic and phenotypic heterogeneity of hypertrophic cardiomyopathy. Int J Mol Med 2016;38:1111-24. [Crossref] [PubMed]
- Maurizi N, Rella V, Fumagalli C, et al. Prevalence of cardiac amyloidosis among adult patients referred to tertiary centres with an initial diagnosis of hypertrophic cardiomyopathy. Int J Cardiol 2020;300:191-5. [Crossref] [PubMed]
- Ruiz-Guerrero L, Barriales-Villa R. Storage diseases with hypertrophic cardiomyopathy phenotype. Glob Cardiol Sci Pract 2018;2018:28. [Crossref] [PubMed]
- Hagège A, Réant P, Habib G, et al. Fabry disease in cardiology practice: Literature review and expert point of view. Arch Cardiovasc Dis 2019;112:278-87. [Crossref] [PubMed]
- Ahluwalia M, Ho CY. Cardiovascular genetics: the role of genetic testing in diagnosis and management of patients with hypertrophic cardiomyopathy. Heart 2021;107:183-9. [Crossref] [PubMed]


