
The transcription factor PPARA mediates SIRT1 regulation of NCOR1 to protect damaged heart cells
Highlight box
Key findings
• This study clarified that Sirtuin 1 (SIRT1) activated the expression of nuclear receptor co-repressor 1 (NCOR1) by up-regulating the transcription factor peroxisome proliferator-activated receptor α (PPARA), thereby alleviating doxorubicin-induced damaged heart cells.
What is known and what is new?
• SIRT1 could reduce heart cells apoptosis and improve cardiac function.
• The regulatory relationship between SIRT1, NCOR1, and PPARA in the progression of cardiac failure remains unclear.
What is the implication, and what should change now?
• The SIRT1/PPARA/NCOR1 pathway play a protective role in damaged heart cells. The study provided a new strategy for heart failure treatment.
IntroductionOther Section
The development of heart failure (HF)
HF is a multifaceted clinical syndrome that develops as a consequence of the progression of diverse heart diseases to an advanced stage (1), characterized by structural or functional damage to the ventricles, leading to symptomatic left ventricular dysfunction (2,3). Patients often present with dyspnea, fluid retention, and significantly shortened survival (4). Despite notable advancements in HF treatment, it remains a prominent cause of cardiovascular morbidity and mortality globally (5), particularly among individuals with pre-existing heart conditions (6,7). To enhance long-term prognoses, mitigate mortality rates, and postpone the onset and progression of HF, further research into potential molecular mechanisms for delaying HF is imperative.
Sirtuin 1 (SIRT1) may play an important role in HF
An abnormal increase in oxidative stress due to severe mitochondrial dysfunction is one of the hallmarks of HF (8). Previous studies showed that SIRT1 participates in and induces the antioxidant expression of superoxide dismutase (SOD) and catalase, thereby reducing oxidative stress and cell apoptosis and protecting the heart from ischemia/reperfusion injury (9,10). SIRT1, in synergistic with sodium-glucose cotransporter 2 (SGLT2) inhibitors, reduces cardiovascular death risk in HF patients (11). These studies suggest that SIRT1 may play an important role in HF.
Peroxisome proliferator-activated receptor α (PPARA) closely associated with cardiac dysfunction
In addition, SIRT1 activates peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) (12,13), which serves as a common transcriptional coactivator of PPARA (14). In a study by Zhu et al., it was observed that cardiomyocyte-specific PPARA deficiency potentiated lipopolysaccharide-induced mitochondrial dysfunction, leading to an increase in cardiac dysfunction (15). Notably, PPARA knockout mice show higher scores of cardiac histological damage than wild-type mice (16). Based on the above research background, it is not difficult to find the potential relationship between SIRT1 and PPARA in HF. Therefore, we aimed to further explore the mechanisms of SIRT1 and PPARA in regulating HF.
Nuclear receptor co-repressor 1 (NCOR1) is an important regulator of myocardial association
Multiple studies have highlighted the significance of PPARA and NCOR1 as key regulators in myocardial function (17,18). Pathological hypertrophy stands as a primary risk factor for HF, with NCOR1 emerging as a potent inhibitor of this detrimental process and associated myocardial dysfunction (19). Li et al. reported that cardiomyocyte-specific NCOR1 knockout mice manifested cardiac hypertrophy at baseline (20). Furthermore, it is reported that the immune effects of NCOR1 are mediated by the suppression of PPAR target genes in both mouse and human macrophages (21). Despite this knowledge, the question of whether the regulation of NCOR1 by PPARA affects the progression of HF remains unexplored.
Based on the background above, we speculated that SIRT1 could protect heart cells and delay the progression of HF through the PPARA/NCOR1 pathway. Thus, we constructed a heart cell injury model in this study to assess the expression of the SIRT1/PPARA/NCOR1 pathway. The SIRT1/PPARA/NCOR1 axis could provide new insights into the molecular mechanisms of HF research.
MethodsOther Section
Cell culture and treatment
Human cardiomyocyte AC16 cell line (AW-CNH103, Abiowell, Changsha, China) were cultured in Dulbecco’s modified eagle medium (C11995500, Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (AWI0070a, Abiowell). Human cardiac microvascular endothelial cell (HCMEC, CP-H079, Procell, Wuhan, China) lines were cultured in a specific medium (AW-MC013, Abiowell). The cultures were incubated at 37 ℃ in a humidified atmosphere with 5% CO2. Logarithmic cells were seeded in 6-well plates and transfected using Lipofectamine 2000 (11668019, Invitrogen, Carlsbad, CA, USA) as described in the kit after cell adhesion. All plasmid sequences were synthesized by HonorGene (Changsha, China).
Heart cell injury was simulated by doxorubicin (DOX, A3966, APExBIO, Houston, TX, USA) intervention AC16 and HCMEC cells. The cells were randomly divided into Control, DOX, overexpression negative control (oe-NC), oe-SIRT1, oe-NC + small interfering RNA negative control (si-NC), oe-SIRT1 + si-NC, oe-SIRT1 + si-PPARA, oe-PPARA + si-NC, and oe-PPARA + si-NCOR1 groups. In the control group, cells were cultured normally and detected after 24 h. In the DOX group, cells were added to 2 µM DOX for 24 h intervention (22). In the oe-NC group, cells were transfected with oe-NC for 24 h before adding 2 µM DOX intervention for 24 h. In the oe-SIRT1 group, cells were transfected with oe-SIRT1 for 24 h before adding 2 µM DOX for 24 h intervention. In the oe-NC + si-NC group, cells were transfected with plasmids of oe-NC and si-NC for 24 h before adding 2 µM DOX for 24 h intervention. In the oe-SIRT1 + si-NC group, cells were transfected with plasmids of oe-SIRT1 and si-NC for 24 h before adding 2 µM DOX for 24 h intervention. In the oe-SIRT1 + si-PPARA group, cells were transfected with plasmids of oe-SIRT1 and si-PPARA for 24 h before adding 2 µM DOX for 24 h intervention. In the oe-PPARA + si-NC group, cells were transfected with plasmids of oe-PPARA and si-NC for 24 h before adding 2 µM DOX for 24 h intervention. In the oe-PPARA + si-NCOR1 group, cells were transfected with plasmids of oe-PPARA and si-NCOR1 for 24 h before adding 2 µM DOX for 24 h intervention.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA extraction was performed following the instructions of the Total RNA Extraction (Trizol, 15596026, Thermo Fisher Scientific, Waltham, MA, USA). The concentration of the extracted RNA was assessed using an ultraviolet spectrophotometer, and its purity was calculated. RNA was reverse-transcribed to cDNA using a reverse transcription kit (CW2569, CWBIO, Taizhou, China). The samples were reacted in a real-time PCR instrument (QuantStudio 1, ABI, Los Angeles, CA, USA) with SYBR Mix. The primer sequences used for SIRT1 amplification were as follows: forward 5'-ATTCCAAGTTCACCCCAT-3' and reverse 5'-TGGCATATTCACCTAACCT-3'. For glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an internal reference gene, the primer sequences were forward 5'-ACAGCCTCAAGATCATCAGC-3' and reverse 5'-GGTCATGAGTCCTTCCACGAT-3'.
Western blot analysis
Cells were collected, and total proteins were extracted with RIPA lysis buffer (AWB0136, Abiowell). The protein concentration was detected using a BCA kit (AWB0156, Abiowell). Subsequently, total proteins were separated by SDS-PAGE and transferred to the nitrocellulose membrane. After the transfer, the membrane was rinsed once in 1×PBST, and then the membrane was completely immersed in the blocking solution and shaken on a shaker for 90 min. SIRT1 (13161-1-AP, 1:3,000, Proteintech, Wuhan, China), myosin heavy chain 6 (Myh6, 22281-1-AP, 1:20,000, Proteintech), myosin heavy chain 6 (Myh7, 22280-1-AP, 1:2,000, Proteintech), GAPDH (10494-1-AP, 1:5,000, Proteintech), PPARA (ab227074, 1:1,000, Abcam, Cambridge, UK), NCOR1 (ab300615, 1:1,000, Abcam) were diluted with 1×PBST in a certain proportion. These primary antibodies were incubated with the membrane overnight at 4 ℃. After rinsing, HRP goat anti-rabbit IgG (SA00001-2, 1:6,000, Proteintech) was incubated with the membrane for 90 min. Finally, the membrane was incubated with enhanced chemiluminescence solution (AWB0005, Abiowell) for 1 min and observed using a chemiluminescence imaging system (ChemiScope 6100, Clinx Science Instruments, Shanghai, China).
Flow cytometry
The Apoptosis Kit (KGA1030, Jiangsu Keygen Biotech Corp., Ltd., China) was used to detect apoptosis. About 3.2×105 cells were collected with ethylene diamine tetraacetic acid (EDTA)-free trypsinization. Then, 5 µL of annexin APC and 5 µL of propidium iodide were incubated with cells at room temperature for 10 min away from light. Finally, the apoptosis rate was detected by flow cytometry (A00-1-1102, Beckman, Brea, CA, USA).
Enzyme-linked immunosorbent assay (ELISA)
Human atrial natriuretic peptide (ANP, CSB-E11193h, CUSABIO, Wuhan, China), human brain natriuretic peptide (BNP, CSB-E07970h, CUSABIO), human interleukin 1β (IL-1β, CSB-E08053h, CUSABIO), human interleukin 18 (IL-18, KE00193, Proteintech) were detected using the ELISA kit. The levels of ANP, BNP, IL-1β, and IL-18 in cells were measured following the instructions.
Reactive oxygen species (ROS), malondialdehyde (MDA), and SOD determination
Cells were collected and prepared into cell suspensions. Following the instructions of the ROS assay kit (E004-1-1, Nanjing Jiancheng Bioengineering Institute, China), cells were incubated with the 2'-7'-dichlorofluorescein diacetate (DCFH-DA) probe for 30–60 min. The mixture was mixed every 3–5 min to allow adequate contact of the probe with the cells. Finally, a fluorescence microscope was used to detect the fluorescence intensity. The excitation and emission wavelengths were set to 500 and 525 nm, respectively.
Cell supernatants were collected to measure the levels of MDA (A003-1, Nanjing Jiancheng Bioengineering Institute) and SOD (A001-3, Nanjing Jiancheng Bioengineering Institute) by assay kits according to the instructions. The absorbance values of the MDA and SOD were measured by a microplate reader (MB-530, HEALES, Shenzhen, China) at 532 and 450 nm, respectively.
Co-immunoprecipitation (Co-IP) analysis
Firstly, AC16 and HCMEC cells were washed with PBS. Then, the cells were lysed with IP cell lysate (AWB0144, Abiowell). Total proteins were obtained by centrifugation of the lysate and subsequently incubated overnight with PPARA (66826-1-Ig, 1:2,000, Proteintech). Twenty microliters of Protein A/G agarose beads were added to capture antigen-antibody complexes, and the antigen-antibody mixture was incubated on a shaker at 4 ℃ for 2 h. After co-immunoprecipitation, the expressions of SIRT1 and PPARA were detected using Western blot.
Determination of adenosine triphosphate (ATP) content
Cells were separated from the culture supernatant through centrifugation, and the cell pellet in the lower layer was collected. A total of 300–500 µL of cold double-distilled water was added to the collected cells and homogenized in an ice bath. Cell ATP contents were measured following the instructions of the ATP content determination kit (A095-1-1, Nanjing Jiancheng Bioengineering Institute). Subsequently, the chromogenic and termination solutions were used and then measured using a spectrometer at a wavelength of 636 nm.
Chromatin immunoprecipitation (ChIP) assay
Following the guidelines in the ChIP Assay Kit (ab500, Abcam) protocol, a final concentration of 1.1% formaldehyde was utilized to crosslink the DNA and proteins in the cells. Subsequently, the crosslinked DNA was fragmented into smaller fragments using ultrasonication. PPARA (#2435, CST, Danvers, MA, USA) or positive control (ab1791, Abcam) were added to the supernatant for immunoprecipitation, respectively. Finally, the eluted and purified DNA as a template amplified the fragment region of the NCOR1 promoter region prediction site by RT-qPCR. The primer sequences were NCOR1-F: ATGTTCGTCAGGCTGGTCTC, NCOR1-R: CGCTTGTAATCCCAGCACTT.
Cell Counting Kit-8 (CCK-8) assay
Cell viability assays were performed using the Cell Counting Kit (AWC0114b, Abiowell). Three complex wells were set up in each group. After adherent culture, 100 µL of medium containing 10% CCK-8 reagent replaced the original medium. Finally, AC16 and HCMEC cells were cultured for 4 h, and then optical density at 450 nm was read by a microplate reader (MB-530, HEALES).
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 8.0 software, and all data were expressed as mean ± standard deviation. If the data followed a normal distribution, we performed pairwise comparisons between the two groups using the t-test. Comparisons between multiple groups were performed using the one-way analysis of variance test, and then Tukey’s multiple comparisons test was used for multiple comparisons. P<0.05 was considered to indicate statistical significance.
ResultsOther Section
SIRT1 protected damaged heart cells
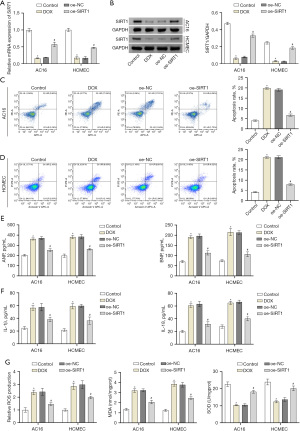
To explore the effects of SIRT1 on AC16 and HCMEC cells, we induced heart cell injury with DOX. SIRT1 expression in AC16 and HCMEC cells was assessed using RT-qPCR and Western blot. The results revealed a significant decrease in SIRT1 expression in the DOX group compared to the control group. At the same time, Western blot verified that oe-SIRT1 and its negative control were successfully transfected (Figure 1A,1B). The results demonstrated a significant increase in the apoptosis rate in the DOX group compared to the control group, but it was significantly reduced after overexpression of SIRT1 (Figure 1C,1D). The contents of HF-related factors and inflammation-related factors in the supernatants of the cell were measured by ELISA, and the results showed that the contents of ANP, BNP, IL-1β and IL-18 were significantly higher in the DOX group than in the control group. Subsequently, the contents of these factors were significantly reduced after overexpression of SIRT1 (Figure 1E,1F). ROS, MDA, and SOD levels were evaluated using the respective assay kits. The results indicated a significant increase in the levels of ROS and MDA in the DOX group compared to the control group, accompanied by a significant decrease in SOD level. When compared to the oe-NC group, the ROS and MDA levels were dramatically reduced, and the SOD content was dramatically increased in the oe-SIRT1 group (Figure 1G). The above results showed that SIRT1 could protect damaged heart cells.
SIRT1 targeted the regulation of PPARA
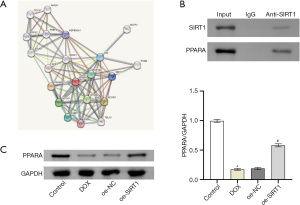
Research showed that PPARA was an important regulatory factor in HF (23,24). The STRING database was used to predict a protein-protein interaction between PPARA and SIRT1 (Figure 2A). First, we verified the relationship between SIRT1 and PPARA by Co-IP assay. The results showed that SIRT1 and PPARA protein were detected in the immune complex obtained with anti-PPARA antibodies, indicating that there was an interaction between SIRT1 and PPARA (Figure 2B). PPARA expression in AC16 and HCMEC cells was assessed by Western blot, and the results indicated that PPARA expression in the DOX group was dramatically lower compared to the control group. However, PPARA expression increased significantly after overexpression of SIRT1 (Figure 2C). The above results showed that SIRT1 targeted the regulation of PPARA.
SIRT1 targeted the regulation of PPARA and affected heart cells
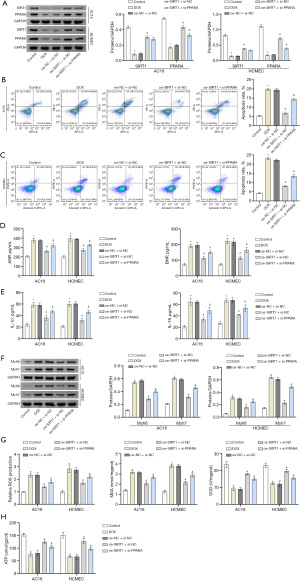
To further explore the effect of SIRT1-targeted regulation of PPARA, we co-transfected AC16 and HCMEC cells with oe-NC, si-NC, oe-SIRT1 or si-PPARA. Western blot analysis indicated that the protein expression of SIRT1 in oe-SIRT1 + si-NC was remarkably higher compared to the oe-NC + si-NC group, but there was no significant difference compared with the oe-SIRT1 + si-PPARA group. The protein expression level of PPARA in the oe-SIRT1 + si-NC group was significantly higher than in the oe-NC + si-NC group and the oe-SIRT1 + si-PPARA group. The above results verified that the transfection was successful (Figure 3A). The apoptosis rate and contents of ANP, BNP, IL-1β and IL-18 were assessed using flow cytometry and ELISA. The results showed that the cell apoptotic rate and the levels of various indicators in the oe-SIRT1 + si-NC group were significantly lower than those in the oe-NC + si-NC group, and they significantly increased after knocking down PPARA (Figures 3B-3E). The expression of the HF marker proteins Myh6 and Myh7 were detected using Western blot, which showed that the expression levels of Myh6 and Myh7 in the oe-SIRT1 + si-NC group decreased significantly compared to the oe-NC + si-NC group. However, after knocking down PPARA, these results were reversed (Figure 3F). At the same time, compared with the oe-NC + si-NC group, ROS and MDA production in the oe-SIRT1 + si-NC group increased significantly. In contrast, SOD activity and ATP content were significantly reduced, and the results were reversed after knocking down PPARA (Figure 3G,3H). Based on the above results, it was not difficult to find that knockdown PPARA could partially reverse the effect of overexpression of SIRT1 on AC16 and HCMEC cells. Therefore, it could be stated that SIRT1 targeted the regulation of PPARA to affect heart cells.
PPARA regulated NCOR1 and protected damaged heart cells
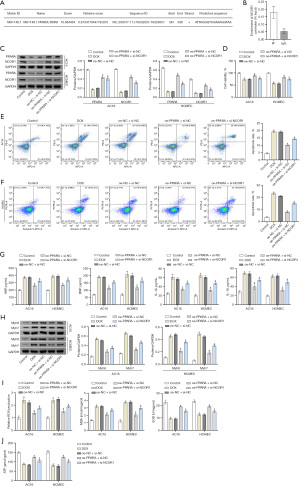
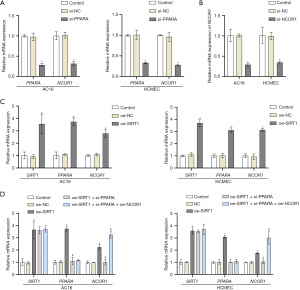
The JASPAR database was used to predict the presence of PPARA binding motifs in the promoter region of NCOR1 (Figure 4A). The enrichment of PPARA in the NCOR1 promoter region was detected using a ChIP assay. The results revealed a significant increase in PPARA enrichment within the NCOR1 promoter region compared to the IgG control group (Figure 4B). Subsequently, the cells were co-transfected with oe-NC, si-NC, oe-PPARA, or si-NCOR1 and then treated with DOX (Figure 4C-4J). The Western blot results revealed a significant increase in PPARA protein expression in the oe-PPARA + si-NC group compared to the oe-NC + si-NC group, with no significant difference compared to the oe-PPARA + si-NCOR1 group. The protein expression of NCOR1 was markedly higher in the oe-PPARA + si-NC group compared to both the oe-NC + si-NC group and oe-PPARA + si-NCOR1 group, indicating successful transfection of the cells (Figure 4C), and the above results showed that the cells were successfully transfected. Moreover, we transfected siRNA to knock down the expression of PPARA and NCOR1 in untreated AC16 and HCMEC cells. As shown in Figure 5A,5B, the knockdown efficiency of PPARA and NCOR1 was around 70%. RT-qPCR results showed a significant decrease in mRNA expression levels of PPARA and NCOR1 in the si-PPARA group compared to the si-NC group (Figure 5A). Then, the cell viability of each group was detected by CCK-8 assay, and the results showed that the cell viability of the oe-PPARA + si-NC group was significantly higher than that of the oe-NC + si-NC group. However, the cell viability was significantly reduced after interfering with NCOR1 (Figure 4D). The results showed that compared with the oe-NC + si-NC group, the apoptosis rate, the contents of ANP, BNP, IL-1β and IL-18, and the protein expression levels of Myh6 and Myh7 were significantly reduced compared with the oe-NC + si-NC group. However, the results were reversed after knocking down NCOR1 expression (Figure 4E-4H). The results of biochemical detection and ATP content detection showed that the ROS and MDA production of the oe-PPARA + si-NC group were significantly reduced compared with the oe-NC + si-NC group. In contrast, the SOD activity and ATP content were significantly increased, and the results were reversed on a one-by-one basis after interfering with NCOR1 (Figure 4I,4J). Based on the above results, knockdown NCOR1 could partially reverse the protective effect of overexpression of PPARA on damaged cells. Hence, it indicated that PPARA could regulate NCOR1 to protect damaged heart cells. Furthermore, overexpression of SIRT1 in untreated cells significantly increased the expression of PPARA and NCOR1 (Figure 5C). In order to better explain the pathway of SIRT1/PPARA/NCOR1, we transfected oe-SIRT1, si-PPARA, and oe-NCOR1 into untreated AC16 and HCMEC cells. As shown in Figure 5D, silencing PPARA reversed the promoting effect of overexpressed SIRT1 on PPARA and NCOR1. Subsequently, overexpression of NCOR1 partially countered the inhibitory effect of silencing PPARA on PPARA and NCOR1. It was worth noting that silencing PPARA or overexpressing NCOR1 did not significantly affect the expression level of SIRT1. In conclusion, our results indicated that SIRT1 activated NCOR1 expression by promoting PPARA expression in AC16 and HCMEC cells.
DiscussionOther Section
HF stands as a prominent contributor to global morbidity and mortality (8,25). Therefore, its molecular mechanism was a research hotspot. To prevent the development of HF, an underlying molecular mechanism needs to be explored. The research on the SIRT1/PPARA/NCOR1 pathway in HF was innovative. In this study, experimental data were obtained through bioinformatics analysis and in vitro cell experiments, and the molecular mechanism of this pathway was elaborated.
Numerous studies demonstrated the involvement of SIRT1 in delaying the progression of HF. Overexpression of SIRT1 could upregulate brain-derived neurotrophic factor (BDNF) and reduce cardiomyocyte apoptosis, thereby improving cardiac function (26). DOX induction could construct HF models in mice and cells, and daidzein exerted antioxidant function and alleviated HF via the SIRT3/Forkhead box O3a (FOXO3a) pathway (27). The expression of SIRT1 was activated in myocardial infarction mice, which could significantly reduce the infarct area and alleviate myocardial infarction-induced apoptosis and HF (28). SIRT1, a class III histone deacetylase (29), could increase endoplasmic reticulum Ca2+-ATPase activity by regulating the acetylation of sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2), improving cardiac function in the case of HF (30). Our study first constructed a cellular HF model by inducing heart cell injury by DOX. Subsequently, we found a low expression of SIRT1 in this model. After overexpression of SIRT1, the apoptosis rate was decreased, the accumulation of ANP, BNP, IL-1β, IL-18, ROS and MAD was reduced, and SOD activity was significantly increased in AC16 and HCMEC cells. These results further demonstrated that SIRT1 protected damaged heart cells and played an important role in alleviating HF.
HF is closely related to mitochondrial biogenesis and function (31,32). Mitochondrial biogenesis is controlled by SIRT1, which interacts with the transcriptional coactivator of PPARγ, thereby increasing oxygen consumption and energy production (24). PPARγ and PPARA are members of the nuclear hormone receptor superfamily (33). Among them, PPARA serves as a selective intracellular fatty acid sensor, playing a pivotal role in regulating lipid remodeling by transactivating a multitude of genes associated with lipid metabolism (34). The expression of PPARA was reduced during HF, leading to decreased fatty acid oxidation and myocardial energy deficiency (24). Chen et al. confirmed PPARA as one of the diagnostic markers related to energy metabolism, which could be used to predict the risk of HF due to myocardial infarction and help optimize clinical treatment (35). These studies implied that PPARA significantly contributes to HF development. Research by Wang et al. indicated that PPARα alleviated angiotensin II-induced hypertensive HF by inhibiting the NF-κB pathway (36). Moreover, activation of the SIRT1-PGCα-PPARα pathway could promote mitochondrial fusion, thereby alleviating diabetes-induced cardiac dysfunction (37). We combined the findings with the bioinformatics analysis that PPARA was a downstream factor of SIRT1, so we guessed that SIRT1 may affect heart cells by targeting and regulating PPARA. In this study, we confirmed the interaction between SIRT1 and PPARA through Co-IP analysis, and overexpression of SIRT1 found that the expression of PPARA protein increased significantly, indicating that PPARA was SIRT1 positively regulated. In addition, we found that knockdown PPARA reversed the protective effect of overexpressing SIRT1 on damaged cells. In conclusion, our study showed that SIRT1 could target and promote the expression of PPARA, thereby alleviating the progression of HF.
The STRING database predicted a potential interaction between the protein PPARA and NCOR1. Our data demonstrated that overexpression of PPARA significantly upregulated the expression of NCOR1, and the ChIP assay verified the binding of PPARA to the promoter region of NCOR1. NCOR1 was demonstrated as a key cardioprotective factor (38,39). Following acute myocardial ischemia/reperfusion injury, there was a dramatic downregulation of NCOR1 (18). In addition, NCOR1 was an important metabolic switch that acted on oxidative metabolic signals (17,40). NCOR1 degradation was associated with increased energy metabolism (41). In this study, AC16 and HCMEC cells apoptosis and oxidative stress were increased after NCOR1 knockdown, and mitochondrial ATP production was reduced, suggesting that mitochondrial function may be hindered. Our results indicated that knocking down NCOR1 reversed the protective effect of overexpression of PPARA on damaged cells. Meanwhile, our study revealed the molecular mechanism by which PPARA targeted regulation of NCOR1 protects injured heart cells, providing a new strategy for treating HF.
However, this study still has some limitations. The results of this study were based on the AC16 and HCMEC cell lines, which might not fully reflect the complex and varied characteristics of HF patients. Therefore, it is necessary to integrate animal experiments and clinical data further to confirm the regulatory role of the SIRT1/PPARA/NCOR1 pathway in the development of HF. Addressing this issue will help enhance the reliability of this study.
ConclusionsOther Section
In conclusion, our study clarified that SIRT1 could target up-regulation of PPARA expression and activate NCOR1 expression through the SIRT1/PPARA/NCOR1 pathway, thereby playing a protective role in injured heart cells. The findings of this study offered a potential novel molecular target for delaying the onset of HF.
AcknowledgmentsOther Section
We thank Grammarly (https://www.grammarly.com/) for assisting language check for this manuscript.
Funding: This study was supported by
FootnoteOther Section
Data Sharing Statement: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-101/dss
Peer Review File: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-101/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-101/coif). M.W. receives fundings from Hunan Provincial Administration of Traditional Chinese Medicine Scientific Research Project (No. 2021118) and University Level Scientific Research Fund Supporting Project of Hunan University of Traditional Chinese Medicine (No. 2019XJJJ058). Y.L. receives fundings from The Collaborative Fund of Hunan University of Chinese Medicine (No. ptkt-99-2024026). Q.Y. receives fundings from Health Commission of Hunan Province (No. 202214055692). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Sridharan S, Kini RM, Richards AM. Venom natriuretic peptides guide the design of heart failure therapeutics. Pharmacol Res 2020;155:104687. [Crossref] [PubMed]
- Golla M, Hajouli S, Ludhwani D. Heart Failure and Ejection Fraction. Treasure Island (FL): StatPearls Publishing; 2024.
- Lv J, Li Y, Shi S, et al. Skeletal muscle mitochondrial remodeling in heart failure: An update on mechanisms and therapeutic opportunities. Biomed Pharmacother 2022;155:113833. [Crossref] [PubMed]
- Chen J, Aronowitz P. Congestive Heart Failure. Med Clin North Am 2022;106:447-58. [Crossref] [PubMed]
- Heidenreich PA, Bozkurt B, Aguilar D, et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2022;145:e876-94. [Crossref] [PubMed]
- Savarese G, Becher PM, Lund LH, et al. Global burden of heart failure: a comprehensive and updated review of epidemiology. Cardiovasc Res 2023;118:3272-87. [Crossref] [PubMed]
- Špinar J, Špinarová L, Vítovec J. Pathophysiology, causes and epidemiology of chronic heart failure. Vnitr Lek 2018;64:834-8. [Crossref] [PubMed]
- Yu H, Zhang F, Yan P, et al. LARP7 Protects Against Heart Failure by Enhancing Mitochondrial Biogenesis. Circulation 2021;143:2007-22. [Crossref] [PubMed]
- Bao M, Huang W, Zhao Y, et al. Verapamil Alleviates Myocardial Ischemia/Reperfusion Injury by Attenuating Oxidative Stress via Activation of SIRT1. Front Pharmacol 2022;13:822640. [Crossref] [PubMed]
- Tian L, Cao W, Yue R, et al. Pretreatment with Tilianin improves mitochondrial energy metabolism and oxidative stress in rats with myocardial ischemia/reperfusion injury via AMPK/SIRT1/PGC-1 alpha signaling pathway. J Pharmacol Sci 2019;139:352-60. [Crossref] [PubMed]
- Packer M. Potential Interactions When Prescribing SGLT2 Inhibitors and Intravenous Iron in Combination in Heart Failure. JACC Heart Fail 2023;11:106-14. [Crossref] [PubMed]
- Salazar G, Cullen A, Huang J, et al. SQSTM1/p62 and PPARGC1A/PGC-1alpha at the interface of autophagy and vascular senescence. Autophagy 2020;16:1092-110. [Crossref] [PubMed]
- Li X, Lozovatsky L, Sukumaran A, et al. NCOA4 is regulated by HIF and mediates mobilization of murine hepatic iron stores after blood loss. Blood 2020;136:2691-702. [Crossref] [PubMed]
- Kalliora C, Kyriazis ID, Oka SI, et al. Dual peroxisome-proliferator-activated-receptor-α/γ activation inhibits SIRT1-PGC1α axis and causes cardiac dysfunction. JCI Insight 2019;5:e129556. [Crossref] [PubMed]
- Zhu XX, Wang X, Jiao SY, et al. Cardiomyocyte peroxisome proliferator-activated receptor α prevents septic cardiomyopathy via improving mitochondrial function. Acta Pharmacol Sin 2023;44:2184-200. [Crossref] [PubMed]
- Standage SW, Waworuntu RL, Delaney MA, et al. Nonhematopoietic Peroxisome Proliferator-Activated Receptor-α Protects Against Cardiac Injury and Enhances Survival in Experimental Polymicrobial Sepsis. Crit Care Med 2016;44:e594-603. [Crossref] [PubMed]
- Lima TI, Valentim RR, Araújo HN, et al. Role of NCoR1 in mitochondrial function and energy metabolism. Cell Biol Int 2018;42:734-41. [Crossref] [PubMed]
- Qin Z, Gao L, Lin G, et al. The nuclear receptor co-repressor 1 is a novel cardioprotective factor against acute myocardial ischemia-reperfusion injury. J Mol Cell Cardiol 2022;166:50-62. [Crossref] [PubMed]
- Grund A, Heineke J. Targeting cardiac hypertrophy through a nuclear co-repressor. EMBO Mol Med 2019;11:e11297. [Crossref] [PubMed]
- Li C, Sun XN, Chen BY, et al. Nuclear receptor corepressor 1 represses cardiac hypertrophy. EMBO Mol Med 2019;11:e9127. [Crossref] [PubMed]
- Oppi S, Nusser-Stein S, Blyszczuk P, et al. Macrophage NCOR1 protects from atherosclerosis by repressing a pro-atherogenic PPARγ signature. Eur Heart J 2020;41:995-1005. [Crossref] [PubMed]
- Zhang JM, Yu RQ, Wu FZ, et al. BMP-2 alleviates heart failure with type 2 diabetes mellitus and doxorubicin-induced AC16 cell injury by inhibiting NLRP3 inflammasome-mediated pyroptosis. Exp Ther Med 2021;22:897. [Crossref] [PubMed]
- Ritterhoff J, McMillen TS, Villet O, et al. Increasing fatty acid oxidation elicits a sex-dependent response in failing mouse hearts. J Mol Cell Cardiol 2021;158:1-10. [Crossref] [PubMed]
- Drosatos K, Pollak NM, Pol CJ, et al. Cardiac Myocyte KLF5 Regulates Ppara Expression and Cardiac Function. Circ Res 2016;118:241-53. [Crossref] [PubMed]
- Obokata M, Sorimachi H, Harada T, et al. Epidemiology, Pathophysiology, Diagnosis, and Therapy of Heart Failure With Preserved Ejection Fraction in Japan. J Card Fail 2023;29:375-88. [Crossref] [PubMed]
- Lin B, Zhao H, Li L, et al. Sirt1 improves heart failure through modulating the NF-κB p65/microRNA-155/BNDF signaling cascade. Aging (Albany NY) 2020;13:14482-98. [Crossref] [PubMed]
- Li H, Zhang M, Wang Y, et al. Daidzein alleviates doxorubicin-induced heart failure via the SIRT3/FOXO3a signaling pathway. Food Funct 2022;13:9576-88. [Crossref] [PubMed]
- Chen L, Li S, Zhu J, et al. Mangiferin prevents myocardial infarction-induced apoptosis and heart failure in mice by activating the Sirt1/FoxO3a pathway. J Cell Mol Med 2021;25:2944-55. [Crossref] [PubMed]
- Shen P, Deng X, Chen Z, et al. SIRT1: A Potential Therapeutic Target in Autoimmune Diseases. Front Immunol 2021;12:779177. [Crossref] [PubMed]
- Gorski PA, Jang SP, Jeong D, et al. Role of SIRT1 in Modulating Acetylation of the Sarco-Endoplasmic Reticulum Ca(2+)-ATPase in Heart Failure. Circ Res 2019;124:e63-80. [Crossref] [PubMed]
- Wu C, Zhang Z, Zhang W, et al. Mitochondrial dysfunction and mitochondrial therapies in heart failure. Pharmacol Res 2022;175:106038. [Crossref] [PubMed]
- Schwartz B, Gjini P, Gopal DM, et al. Inefficient Batteries in Heart Failure: Metabolic Bottlenecks Disrupting the Mitochondrial Ecosystem. JACC Basic Transl Sci 2022;7:1161-79. [Crossref] [PubMed]
- Christofides A, Konstantinidou E, Jani C, et al. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism 2021;114:154338. [Crossref] [PubMed]
- Lin Z, Liu J, Long F, et al. The lipid flippase SLC47A1 blocks metabolic vulnerability to ferroptosis. Nat Commun 2022;13:7965. [Crossref] [PubMed]
- Chen H, Jiang R, Huang W, et al. Identification of energy metabolism-related biomarkers for risk prediction of heart failure patients using random forest algorithm. Front Cardiovasc Med 2022;9:993142. [Crossref] [PubMed]
- Wang M, Luo W, Yu T, et al. Corynoline protects ang II-induced hypertensive heart failure by increasing PPARα and Inhibiting NF-κB pathway. Biomed Pharmacother 2022;150:113075. [Crossref] [PubMed]
- Hu L, Guo Y, Song L, et al. Nicotinamide riboside promotes Mfn2-mediated mitochondrial fusion in diabetic hearts through the SIRT1-PGC1α-PPARα pathway. Free Radic Biol Med 2022;183:75-88. [Crossref] [PubMed]
- Metra M, Teerlink JR. Heart failure. Lancet 2017;390:1981-95. [Crossref] [PubMed]
- Geiger MA, Guillaumon AT, Paneni F, et al. Role of the Nuclear Receptor Corepressor 1 (NCOR1) in Atherosclerosis and Associated Immunometabolic Diseases. Front Immunol 2020;11:569358. [Crossref] [PubMed]
- Paluvai H, Shanmukha KD, Tyedmers J, et al. Insights into the function of HDAC3 and NCoR1/NCoR2 co-repressor complex in metabolic diseases. Front Mol Biosci 2023;10:1190094. [Crossref] [PubMed]
- Oliveira AG, Oliveira LD, Cruz MV, et al. Interaction between poly(A)-binding protein PABPC4 and nuclear receptor corepressor NCoR1 modulates a metabolic stress response. J Biol Chem 2023;299:104702. [Crossref] [PubMed]


