
Artificial intelligence machine learning based evaluation of elevated left ventricular end-diastolic pressure: a Cleveland Clinic cohort study
Highlight box
Key findings
• Machine learning (ML) approaches predict elevated left ventricular end-diastolic pressure (LVEDP) and tau with high accuracy, allowing for increased clinical utility of echocardiographic data and interpretation of diastology.
What is known and what is new?
• LVEDP is a useful marker in many cardiac conditions, but requires invasive methods for accurate determination.
• We demonstrate that ML approaches can make use of non-invasive echocardiographic parameters to estimate LVEDP with accuracy.
What is the implication, and what should change now?
• ML approaches present novel ways to integrate echocardiographic, biomarker, and clinical data for cardiovascular evaluation and diastolic function assessment.
Introduction
Background
Left ventricular end-diastolic pressure (LVEDP) is an indicator of diastolic dysfunction, and thus it is an important clinical parameter. Diastolic dysfunction is a key index in determining cardiovascular health, as it is present in many cardiovascular conditions, such as hypertensive heart disease and left ventricular (LV) hypertrophy, and is associated with increased mortality (1,2). The current gold-standard method of measuring LVEDP is intra-cardiac catheterization, which is invasive. The current American Society of Echocardiography (ASE) diastology guidelines draw on a combination of echocardiographic parameters, including early (E) and late (A) mitral diastolic inflow peak velocities, peak early diastolic velocities of both the septal and lateral mitral annulus (e'), peak tricuspid regurgitation (TR) velocity, left atrial volume index (LAVi) to classify patients as indeterminate, normal, or having grade I, II, or III diastolic dysfunction (3).
Rationale and knowledge gap
Although the current ASE guidelines for diagnosing and classifying diastolic dysfunction are widely applied clinically, the individual echocardiographic parameters making up the diagnostic criteria show at best moderate correlation with LVEDP (3). Specifically, the associations of variables such as E/e', LAVi, and TR velocity with LVEDP are confounded by comorbidities including pulmonary hypertension, arrhythmias, tachycardia, and other valvular pathologies (3,4). Therefore, non-invasive correlates of elevated LVEDP are not consistently accurate (2). Moreover, up to 29% of patients are classified by the 2016 ASE guidelines as indeterminate, showing yet another significant shortcoming in conventional assessment (5).
Machine learning (ML) uses algorithms to model patterns in data, and it has the ability to identify relationships that may be too heterogeneous to capture using traditional statistics and knowledge of physicians (6,7). Due to its potential to increase the accessibility and accuracy of diagnosis and prognosis, ML has an expanding role in diastology research (6-8). Previous studies have showed that ML can be promising in its ability to grade diastolic dysfunction (4,9,10). However, there has been little research examining the role of ML to detect elevated LVEDP, which is a widely utilized clinical parameter guiding clinicians in important therapy decisions.
Objective
Therefore, in this study, we primarily assessed the ability of ML models to detect elevated LVEDP in cardiac patients. Our secondary focus was to predict the presence of elevated tau, an invasive measure of LV relaxation time which is considered to be the gold standard of diastolic function (11). Finally, we aimed to evaluate the ability of ML models to distinguish between clinical diagnoses including aortic stenosis (AS), coronary artery disease (CAD), LV hypertrophy (LVH), and left ventricle dysfunction (LVD), using clinical, echocardiographic, and biomarker parameters obtained from non-invasive procedures. This study offers tools for multiple clinical applications. Most importantly, ML models could be used as a non-invasive screening tool to identify elevated LVEDP and tau and ensure prompt treatment in asymptomatic populations. In higher-risk populations, ML models could be used as valuable diagnostic aids. We present this article in accordance with the TRIPOD+AI reporting checklist (available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-128/rc).
Methods
Study population and data pre-processing
The protocol for the present study was approved by the Institutional Review Board of the Cleveland Clinic (IRB: 19-803) and was compliant with the Health Insurance Portability and Accountability Act regulations. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The requirement for individual consent was waived as part of the approved IRB protocol, because identifying information was not used in the analysis and writing of this work. We constructed a retrospective cohort of 460 consecutive patients undergoing echocardiography and left heart catheterization (within 24 hours) without a history of atrial fibrillation and significant mitral valve disease between January 2008 and October 2010 from the Cleveland Clinic (3). Initially, 1,204 patients were identified. Patients with insufficient and inadequate echocardiographic data (n=329), patients with abnormal cardiac rhythm at time of the studies including atrial fibrillation and tachyarrhythmias (n=82), patients with cardiac structural abnormalities including heart transplant, prior mitral valve surgery, mitral stenosis and mitral annular calcification, severe mitral and aortic regurgitation (n=323), and patients with changes in diuretic or vasodilator medication therapy changes between cardiac catheterization (n=10) were excluded. These patients were diagnosed with CAD (n=247), LVD (n=99), LVH (n=56), AS (n=51), and TR (n=7). CAD was defined as stenosis of 50% or more in at least one epicardial coronary artery, while LVH was defined as an LV mass index greater than 115 g/m2 in men and 95 g/m2 in women (12). AS was defined according to current guidelines, with severe AS fulfilling the following criteria: aortic valve area less than 1.0 cm² and a mean transaortic gradient ≥40 mmHg, or a peak transaortic velocity ≥4.0 m/s (13). We were also able to calculate tau (11,14), an invasive measure of LV relaxation time, for 400 of these patients. We included a variety of clinical, echocardiographic, and biomarker parameters. Clinical features included variables such as sex, self-reported race, and age. A variety of echocardiographic parameters were used, including key parameters such as LV ejection fraction, right ventricular systolic pressure, left atrium volume, E/A ratio, mitral valve deceleration time, medial mitral annulus early diastolic velocity (septal e'), lateral mitral annulus early diastolic velocity (lateral e'), and E/e'. N-terminal brain natriuretic peptide (NT-proBNP) was included as a biomarker feature. To reduce multicollinearity in the features, we conducted variance inflation factor (VIF) analysis and removed features with high VIFs (VIF >10). A full list of features can be found in Table S1.
Classifier development
In this study, we performed the following ML experiments. First, we investigated whether a combination of clinical, echocardiographic, and biomarker features can be used to predict elevated LVEDP. Second, we created binary-class classification models to predict elevated tau, an invasively derived time constant of LV relaxation which is considered to be the gold-standard index of LV diastolic function (11). Cut-offs were applied to the numerical LVEDP and tau variables to convert them to classification problems. LVEDP was transformed into a binary categorical outcome of >20 and ≤20 mmHg. Tau was transformed into a binary categorical outcome of >45 and ≤45 ms (11). Finally, we built multi-class classification models to classify the cardiac conditions of CAD, LVH, LVD, and AS.
For all three experiments, the algorithms used were logistic regression (LR), random forest (RF), gradient boosting (GB), support vector machine (SVM) (with radial basis function kernel), and K-nearest neighbors (KNN). Figure S1 shows the workflow for the classification analysis. The dataset was randomly split using a 9:1 ratio into training and test sets. To find the optimal hyperparameters, the training data was further randomly split using a 9:1 ratio into training and validation sets. The entire process was repeated for 20 iterations. Missing data was imputed based on the training set using mean values and continuous data points were standardized. We performed a grid search using each combination of the hyperparameters in Table S2. We selected the set of hyperparameters producing the largest area under the receiver operating characteristic curve (AUROC) for the final models. For the multi-class classification, a one-vs-rest (OvR) strategy was employed, meaning one diagnosis was taken as a positive case, and the remaining diagnoses were taken as the negative case. The models were then trained on the entire training set and tested on the test set. AUROC was used to assess the final model performances.
Interpretation of the ML models
We sought to understand which features were influential in the ML prediction to better understand what variables might be relevant in a clinical setting. To do this, we analyzed the LR algorithms used in this study, as LR was among the top performing models across the tasks. Model weights were extracted from the final trained LR model of each of the 20 iterations, and we calculated the absolute coefficient of variation and the relative absolute value weight as below:
where is the weight for feature , is the entire feature set, and is the sign function. Using a previously defined method, but modifying the cutoff values due to having a smaller patient sample size, features whose absolute coefficient of variation was less than 1 and whose relative weight was greater than 0.2 were considered clinically relevant variables (15).
Statistical analysis
Two-sample t-test for continuous variables and Chi-squared test for categorical variables were conducted for the cohort analysis. Mann-Whitney U-test was used for variables that were not normally distributed, and Fisher’s exact test were used for variables with low frequencies. All the P values calculated were two-sided. The Shapiro-Wilk test was used to perform test of normality. Continuous variables were reported as mean ± standard deviation (SD) if normally distributed and as the median and interquartile range (IQR) if not normally distributed. Categorical data were expressed as frequency (percentage).
All analyses were performed using Python 3.9.12, and all classification models were developed using the package scikit-learn 1.0.2 (16). Code for training and evaluation processes were from https://github.com/ChengF-Lab/CO-ML.
Results
The final study population included 460 patients whose characteristics are summarized in Table 1. The median age was 63 years (IQR, 55–73 years) with a mean body mass index (BMI) of 28.88 kg/m2 (SD =5.79 kg/m2). The majority of patients were male (61.1%) and identified as White (77%). Patients were diagnosed with CAD (n=247), LVD (n=99), LVH (n=56), and AS (n=51), and TR (n=7). Due to the low number of cases for TR, we did not build classification models for this condition. The average LVEDP was 16.04 mmHg (SD =7.41 mmHg), with 88 (19.1%) patients having LVEDP >20 mmHg.
Table 1
| Variables | Overall | LVEDP | Tau | |||||
|---|---|---|---|---|---|---|---|---|
| ≤20 mmHg | >20 mmHg | P value | ≤45 ms | >45 ms | P value | |||
| N | 460 (unless otherwise specified) | 372 | 88 | 178 | 222 | |||
| Age, median [Q1, Q3] (years) | 63.00 [55.00, 73.00] | 63.00 [54.00, 72.00] | 63.50 [58.00, 74.25] | 0.26a | 63.00 [52.00, 72.00] | 63.00 [56.00, 73.00] | 0.34a | |
| Sex: male, n (%) | 281 (61.1) | 231 (62.1) | 50 (56.8) | 0.43 | 102 (57.3) | 141 (63.5) | 0.25 | |
| Race: White, n (%) | 354 (77.0) | 292 (78.5) | 62 (70.5) | 0.14 | 139 (78.1) | 169 (76.1) | 0.73 | |
| BMI, mean (SD) (kg/m2) | 28.88 (5.79) | 28.17 (5.31) | 31.88 (6.71) | <0.001 | 28.39 (5.98) | 29.13 (5.79) | 0.22 | |
| Hypertension, n (%) | 328 (71.3) | 262 (70.4) | 66 (75.0) | 0.35 | 133 (76.0) | 150 (68.2) | 0.11 | |
| IVRT, mean (SD) (ms) | 104.37 (24.31) | 106.65 (23.65) | 94.68 (24.84) | <0.001 | 89.72 (18.37) | 116.53 (22.40) | <0.001 | |
| E/e', mean (SD) | 11.67 (5.32) | 11.12 (4.93) | 14.00 (6.24) | <0.001 | 11.28 (5.34) | 12.00 (5.14) | 0.17 | |
| Ln BNP, mean (SD) (pg/mL) | 4.62 (1.49) | 4.38 (1.44) | 5.45 (1.33) | <0.001 | 4.43 (1.49) | 4.75 (1.47) | 0.13 | |
| Cardiovascular condition, n (%) | <0.001b | <0.001b | ||||||
| Tricuspid regurgitation | 7 (1.5) | 5 (1.3) | 2 (2.3) | 5 (2.8) | 2 (0.9) | |||
| Coronary artery disease | 247 (53.7) | 214 (57.5) | 33 (37.5) | 117 (65.7) | 99 (44.6) | |||
| Left ventricular hypertrophy | 56 (12.2) | 46 (12.4) | 10 (11.4) | 14 (7.9) | 33 (14.9) | |||
| Left ventricle dysfunction | 99 (21.5) | 71 (19.1) | 28 (31.8) | 24 (13.5) | 61 (27.5) | |||
| Aortic stenosis | 51 (11.1) | 36 (9.7) | 15 (17.0) | 18 (10.1) | 27 (12.2) | |||
| LVEDP, mean (SD) (mmHg) | 16.04 (7.41) | – | – | – | 12.76 (6.12) | 18.24 (7.09) | <0.001 | |
| Elevated LVEDP, n (%) | 88 (19.1) | – | – | – | 15 (8.4) | 59 (26.6) | <0.001 | |
| Tau, mean (SD) (ms) | 48.80 (14.18) | 46.49 (12.46) | 58.98 (16.68) | <0.001 | – | – | – | |
| Elevated tau, n (%) | 222 (55.5) | 163 (50.0) | 59 (79.7) | <0.001 | – | – | – | |
Statistical testing was done using a two-sample t-test for continuous variables and Chi-squared test for categorical variables unless otherwise noted. a, Mann-Whitney U-test; b, Fisher’s exact test. Elevated LVEDP and elevated tau were defined as >20 mmHg and >45 ms, respectively. Note: (I) not all subjects had accurate BP readings at the time; (II) there were 400 patients with Tau measurements. LVEDP, left ventricular end-diastolic pressure; BMI, body mass index; SD, standard deviation; IVRT, isovolumic relaxation time; Ln BNP, Ln brain natriuretic peptide; BP, blood pressure.
LVEDP classification
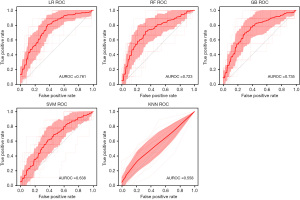
Elevated LVEDP is an important parameter that influences clinical decision-making. To assess if ML can be used to predict elevated LVEDP, we performed binary classification of LVEDP using the LR, RF, GB, SVM, and KNN algorithms. The average AUROC across 20 iterations was 0.761 for LR [95% confidence interval (CI): 0.725–0.796], 0.723 for RF (95% CI: 0.676–0.769), 0.735 for GB (95% CI: 0.692–0.779), 0.638 for SVM (95% CI: 0.593–0.683), and 0.558 for KNN (95% CI: 0.512–0.603) (Figure 1).
Tau and cardiac condition classification
Figure 2 shows the classification results for tau. The average AUROC across 20 iterations was 0.832 for GB (95% CI: 0.700–0.964), 0.822 for LR (95% CI: 0.702–0.941), 0.816 for RF (95% CI: 0.658–0.974), 0.754 for SVM (95% CI: 0.621–0.887), and 0.617 for KNN (95% CI: 0.435–0.799).
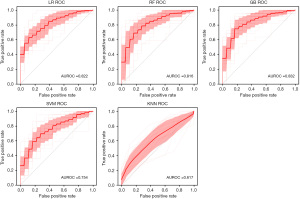
Figure S2 shows the average OvR AUROCs for the diagnoses of CAD, LVD, LVH, and AS. The ranges for average OvR AUROCs were 0.804–0.938 for LR, 0.757–0.975 for RF, 0.787–0.969 for GB, 0.719–0.912 for SVM, and 0.593–0.713 for KNN. The top performing models for the four outcomes have AUROCs of 0.926 for CAD, 0.827 for LVH, 0.975 for LVD, and 0.822 for AS.
Model interpretation
We sought to understand which features are predictive for the outcomes. Since LR generally was among the top performing models across all the tasks, we used the LR models to evaluate the features (Figure 3). For the analysis of elevated LVEDP, a combination of clinical variables (diastolic blood pressure, BMI, and heart rate), echocardiographic variables (such as pulmonary artery systolic pressure, left atrial volume, color M-mode propagation velocity, mitral valve deceleration time, E wave duration, A wave velocity, A' septal, A' lateral, E/e'), and one biomarker variable, NT-proBNP, were influential to the algorithm’s prediction process. For the prediction of elevated tau, variables such as left ventricle ejection fraction, left ventricle mass index, isovolumetric relaxation time, A' septal, average S' integral, and E' lateral were influential for the prediction process.

Interpretability analysis for the models that predicted elevated tau show several shared clinically relevant variables to the LVEDP analysis, including pulmonary artery acceleration time, isovolumetric relaxation time, A' septal, and NT-proBNP. A number of clinical and echocardiographic variables were also clinically relevant for the multi-class classification of the cardiac conditions (Figure S3).
Discussion
Key findings
Elevated LVEDP is associated with advanced diastolic dysfunction, which is an indicator of many cardiovascular diseases. Diastolic dysfunction can be assessed non-invasively using echocardiographic imaging, however, echocardiographic parameters have been only weakly associated with LVEDP due to various confounding comorbidities. This study trains ML algorithms to use echocardiographic features, as well as clinical and biomarker features, to predict elevated LVEDP (>20 mmHg). We also created models to predict elevated tau (>45 ms) and classify various cardiac diagnoses, including AS, CAD, LVH, and LVD.
Explanations of findings
We find that ML algorithms are able to use echocardiographic, clinical, and biomarker features to predict binary classification of LVEDP >20 mmHg. For this analysis, LR was the best performing algorithm (AUROC =0.761), and KNN was the worst (AUROC =0.558). These comprehensive evaluations reveal potential clinical applications of these ML models for assistant of patient care. LR has the additional advantage of being interpretable. The weight analysis of the LR models revealed that variables such as diastolic blood pressure, BMI, and heart rate, pulmonary artery systolic pressure, pulmonary artery acceleration, mitral valve A velocity, mitral valve deceleration time, isovolumetric relaxation time, color M-mode propagation velocity, and NT-proBNP are most influential on the model’s prediction.
Comparison with similar research
There is literature supporting an association between LVEDP and the clinical variables we found, such as BMI (17). Previous studies using traditional statistics have shown poor correlation between LVEDP and echocardiographic variables (3), and non-invasive correlates of elevated LVEDP are not consistently accurate (2). Our ML models’ reliance on weighted echocardiographic variables to predict LVEDP may indicate that these algorithms may be able to detect highly heterogeneous, nuanced relationships in the data that traditional statistics struggles at. These variables may therefore be worthwhile to consider in clinical practice when evaluating patients’ risk for elevated LVEDP. For example, we could build risk calculators by implementing these predictive ML models and clinically actionable variables for risk assessment of LVEDP. However, further clinical validations using independent patient cohorts are warranted before clinical use in the future. ML can also be used to predict the presence of elevated tau, with AUROC as high as 0.832 for the GB algorithm. Finally, we found that ML can be used to do multi-class classification to distinguish the cardiovascular diagnoses, with AUROCs of 0.926 for CAD, 0.827 for LVH, 0.975 for LVD, and 0.822 for AS. Although LR did not have the highest AUROCs for some of these predictions, LR is consistently among the top performing algorithms and offers more interpretable models. In addition, we found that LR showed stable performances for different subgroups of patients by race, sex, or cardiac condition (Figures S4-S6). These results prompted us to further investigate the variables using the LR models.
Previous ML studies have been done to non-invasively predict elevated LVEDP, using orthogonal voltage gradient and photoplethysmography data (18,19). This current study uses a combination of clinical, echocardiographic, and biomarker data. To the best of our knowledge, this study is the first ML study to predict elevated tau. Although tau is less commonly discussed than LVEDP, tau is considered the gold standard for evaluation of diastolic function, and is a more accurate marker of diastolic function compared to LVEDP alone (1,20). Tau is not routinely used clinically, as it is difficult to calculate, and precise measurement of tau requires high-fidelity invasive catheters. Age and sex have previously been shown to be useful clinically in predicting CAD; however, these variables did not show strong predictive performance in this study (21). A potential explanation for this observation is that inclusion of the more specific echocardiographic and biomarker parameters in this study may have overshadowed the influence of these two clinical variables. In addition, as the models were trained using the OvR strategy, the clinically relevant variables we discovered are therefore associated with distinguishing the cardiac conditions. In summary, our comprehensive studies demonstrated the clinical utilities of predictive ML models in assessment of LVEDP when we leveraged clinical, echocardiographic, and biomarker data from individuals.
Strengths and limitations
Our study has several strengths and limitations. First, all patient data were manually curated and checked to ensure high quality. Importantly, we were able to obtain tau, which is considered the gold standard of diastolic function, for 87% patients (n=400). Also unique to our study is the inclusion of a sizable cohort of patients with both detailed echocardiographic and invasive hemodynamic variables. However, we acknowledge that our study has a relatively small sample size of patients for several cardiac conditions (Table 1). For example, there were only seven cases of TR, and therefore this diagnosis was excluded from the multi-class classification. The small sample sizes of some of the conditions may result in large false negative errors. In addition, 77% of the patients were White, which could potentially limit model generalizability to other racial groups. The median age was 63 years, representing an older patient demographic. Our dataset also has limited data regarding additional comorbidities and there may be variables absent in our dataset that may confound the predictions.
To improve the interpretability of our models, we conducted VIF analysis and refined the feature set to reduce multicollinearity. Although the final feature set used throughout this study has low VIF values (Figure S7), we found that the performance could be slightly improved with more features (Figure S8). Overfitting is a potential issue of training ML models. To address this, the grid search process helped select hyperparameters that would limit overfitting during the training and validation process. For example, L2 regularization was applied for LR, and limitations to tree depth and subsampling were applied for GB. Subgroup analyses (Figures S4-S6) showed that LR generalizes well in different smaller subgroups with similar performances to the entire cohort, compared to other ML algorithms.
Implications and actions needed
In future work, external validation using data from independent patient cohorts or cohorts from different healthcare systems would help increase validity of these results. In addition, other ML methods, such as neural networks, could also be applied to increase prediction performance. Raw image data, including echocardiographic images could also be obtained to increase the number of features available for model training and prediction in the future.
Conclusions
In conclusion, we demonstrate that ML algorithms can robustly predict the presence of elevated LVEDP (>20 mmHg) and elevated tau (>45 ms). ML could be used to aid the clinical interpretation of echocardiographic data, and provide a way to integrate echocardiographic, clinical, and biomarker data for improved cardiovascular assessment of intra-cardiac pressures.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD+AI reporting checklist. Available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-128/rc
Data Sharing Statement: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-128/dss
Peer Review File: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-128/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (https://cdt.amegroups.com/article/view/10.21037/cdt-24-128/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The protocol for the present study was approved by the Institutional Review Board of the Cleveland Clinic (IRB: 19-803). The requirement for individual consent was waived as part of the approved IRB protocol, because identifying information was not used in the analysis and writing of this work.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nagueh SF. Left Ventricular Diastolic Function: Understanding Pathophysiology, Diagnosis, and Prognosis With Echocardiography. JACC Cardiovasc Imaging 2020;13:228-44. [Crossref] [PubMed]
- Playford D, Strange G, Celermajer DS, et al. Diastolic dysfunction and mortality in 436 360 men and women: the National Echo Database Australia (NEDA). Eur Heart J Cardiovasc Imaging 2021;22:505-15. [Crossref] [PubMed]
- Sato K, Grant ADM, Negishi K, et al. Reliability of updated left ventricular diastolic function recommendations in predicting elevated left ventricular filling pressure and prognosis. Am Heart J 2017;189:28-39. [Crossref] [PubMed]
- Lancaster MC, Salem Omar AM, Narula S, et al. Phenotypic Clustering of Left Ventricular Diastolic Function Parameters: Patterns and Prognostic Relevance. JACC Cardiovasc Imaging 2019;12:1149-61. [Crossref] [PubMed]
- Almeida JG, Fontes-Carvalho R, Sampaio F, et al. Impact of the 2016 ASE/EACVI recommendations on the prevalence of diastolic dysfunction in the general population. Eur Heart J Cardiovasc Imaging 2018;19:380-6. [Crossref] [PubMed]
- Haq IU, Haq I, Xu B. Artificial intelligence in personalized cardiovascular medicine and cardiovascular imaging. Cardiovasc Diagn Ther 2021;11:911-23. [Crossref] [PubMed]
- Haq IU, Chhatwal K, Sanaka K, et al. Artificial Intelligence in Cardiovascular Medicine: Current Insights and Future Prospects. Vasc Health Risk Manag 2022;18:517-28. [Crossref] [PubMed]
- Xu B, Kocyigit D, Grimm R, et al. Applications of artificial intelligence in multimodality cardiovascular imaging: A state-of-the-art review. Prog Cardiovasc Dis 2020;63:367-76. [Crossref] [PubMed]
- Pandey A, Kagiyama N, Yanamala N, et al. Deep-Learning Models for the Echocardiographic Assessment of Diastolic Dysfunction. JACC Cardiovasc Imaging 2021;14:1887-900. [Crossref] [PubMed]
- Tromp J, Seekings PJ, Hung CL, et al. Automated interpretation of systolic and diastolic function on the echocardiogram: a multicohort study. Lancet Digit Health 2022;4:e46-54. [Crossref] [PubMed]
- Senzaki H, Fetics B, Chen CH, et al. Comparison of ventricular pressure relaxation assessments in human heart failure: quantitative influence on load and drug sensitivity analysis. J Am Coll Cardiol 1999;34:1529-36. [Crossref] [PubMed]
- Lang RM, Badano LP, Mor-Avi V, et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr 2015;28:1-39.e14. [Crossref] [PubMed]
- Otto CM, Nishimura RA, Bonow RO, et al. 2020 ACC/AHA Guideline for the Management of Patients With Valvular Heart Disease: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2021;143:e35-71. [Crossref] [PubMed]
- Scalia GM, Greenberg NL, McCarthy PM, et al. Noninvasive assessment of the ventricular relaxation time constant (tau) in humans by Doppler echocardiography. Circulation 1997;95:151-5. [Crossref] [PubMed]
- Zhou Y, Hou Y, Hussain M, et al. Machine Learning-Based Risk Assessment for Cancer Therapy-Related Cardiac Dysfunction in 4300 Longitudinal Oncology Patients. J Am Heart Assoc 2020;9:e019628. [Crossref] [PubMed]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825-30.
- Russo C, Jin Z, Homma S, et al. Effect of obesity and overweight on left ventricular diastolic function: a community-based study in an elderly cohort. J Am Coll Cardiol 2011;57:1368-74. [Crossref] [PubMed]
- Bhavnani SP, Khedraki R, Cohoon TJ, et al. Multicenter validation of a machine learning phase space electro-mechanical pulse wave analysis to predict elevated left ventricular end diastolic pressure at the point-of-care. PLoS One 2022;17:e0277300. [Crossref] [PubMed]
- Fathieh F, Paak M, Khosousi A, et al. Predicting cardiac disease from interactions of simultaneously-acquired hemodynamic and cardiac signals. Comput Methods Programs Biomed 2021;202:105970. [Crossref] [PubMed]
- Grant AD, Negishi K, Negishi T, et al. Grading diastolic function by echocardiography: hemodynamic validation of existing guidelines. Cardiovasc Ultrasound 2015;13:28. [Crossref] [PubMed]
- Goff DC Jr, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 2014;63:2935-59. [Crossref] [PubMed]


