
Actionable secondary findings in arrhythmogenic right ventricle cardiomyopathy genes: impact and challenge of genetic counseling
Introduction
The rapid evolution and widespread use of high throughput sequencing in clinical laboratories has allowed an incredible progress in the genetic diagnostics of several inherited disorders. However, the new technologies have brought new challenges. Like any test or procedure, comprehensive sequencing analyses are able to detect findings for conditions beside the primary diagnostic request. These findings are so-called secondary findings (SF), variants in genes not directly related to the primary clinical diagnosis. The American College of Medical Genetics and Genomics (ACMG) has recommended the report of SF in 59 clinically actionable genes (ACMG v2.0) (1,2), including 27 genes linked to cardiovascular phenotypes such as Marfan's syndrome (MFS), vascular Ehlers-Danlos syndrome (EDS type IV), Loeys-Dietz syndrome (LDS), familial thoracic aortic aneurysm/dissection (fTAAD), catecholaminergic polymorphic ventricular tachycardia (CPVT), long QT syndrome (LQTS), Brugada syndrome (BrS), Fabry's disease, familial hypertrophic and dilated cardiomyopathy (HCM, and DCM), left ventricle non-compaction cardiomyopathy (LVNC) and arrhythmogenic right ventricular cardiomyopathy (ARVC). Since the hereditary conditions related to the actionable genes show incomplete penetrance and variable expressivity, it is possible that individuals with pathogenic variants in these genes will never develop symptoms. Considerable literature discusses the utility and ethics of reporting SF (3,4). Some have argued that SF in diagnostic sequencing should not be reported at all until there is strong evidence of benefit, while others have advocated that variations all disease-associated genes could be medically useful and should be reported (5). The ACMG appointed a Working Group on SF in comprehensive NGS analyses to make recommendations dealing with the management of SF (6). Depending on the gene, the SF Group recommends that laboratories report only the types of variants within the actionable genes that have been previously reported as disease causing (known pathogenic, KP) and variants that are previously unreported but are of the type which is expected to cause the disorder (expected pathogenic, EP) (6).
One of the selected condition represented in the ACMG actionable gene list is ARVC, a rare inherited heart-muscle disorder (typically autosomal-dominant) that causes progressive fibro‐fatty replacement of (mainly) the right ventricular myocardium, predisposing affected individuals to ventricular arrhythmia and SCD (7,8). Because of the clinical and genetic heterogeneity of ARCV, SCD may be the first clinical manifestation, clinical management of asymptomatic individuals with variants in ARVC genes is challenging. The prevalence of ARVC is estimated to be 1:1,000–1:1,250 (9-11) in the general population, and accounts for up to 22% of SCD cases among young adults (<35 years) and athletes (12-15).
The diagnosis is based on the revised 2010 Task Force Criteria (16), and is a combination of major and minor criteria from different areas including right ventricle function and structure, electrocardiogram (ECG) findings as well as genetic or familial background. Different from all other forms of cardiomyopathy, the Task Force Criteria for ARVC include the presence of a pathogenic variant in ARVC-related genes as a major criterion to establish the diagnosis (16). Variants in desmosomal genes accounting for 40–50% of cases (17). The majority of the detected causal variants being located in PKP2 (45–73% of all causal variants) which encodes Plakophilin-2 (18). Other desmosomal genes include DSC2 (Desmocollin-2) (19), DSG2 (Desmoglein-2) (20), DSP (Desmoplakin) (21) and JUP (Junction-Plakoglobin) (22), which cause the disease mainly in the autosomal-dominant mode, apart for JUP, which causes ARVC in an autosomal recessive mode. For DSP, DSC2 and more recently DSG2 autosomal-recessive ARVC cases have been rarely described (23,24). Several non-desmosomal genes have also been casually implicated in ARVC pathogenesis, including variants in TMEM43 (25) (encoding for the transmembrane protein TMEM43) and PLN (phospholamban) (26). The ACMG recommended the return of pathogenic variants in five of these ARVC genes: DSC2, DSG2, DSP, PKP2 and TMEM43. Importantly, variants are less likely to be classified as pathogenic in the absence of clinical symptoms or family history, whereas they are more likely pathogenic in a symptomatic individual (27,28). However, there are limited data on the frequency of such pathogenic variants, and a standardized level of evidence for pathogenicity assessment of these variants has not been defined.
No causal therapy is available for ARVC, so that the focus of current management is on early identification of asymptomatic patients at risk through molecular diagnostic and clinical cascade screening of family members, optimal SCD risk stratification, and timely initiation of preventative therapies to avoid disease progression. Several studies emphasized the challenge of managing SF in ARVC-related genes (27,29,30). A high frequency of pathogenic variants in genes associated with ARVC in population-based control cohorts indicate that a proportion of putative ARVC-causing variants may be inaccurately classified (29).
Here, we examined the frequency of SF in genes linked to inherited CVD in 6605 individuals, who underwent genetic testing for noncardiac reason and no documented incidence of CVD. In particular, we focused on the pathogenicity assessment of the identified variants in actionable ARVC genes. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/cdt-20-585).
Method
Patient cohort
6605 NGS data were analysed for variants in selected actionable genes of the ACMG secondary findings (SF) v2.0 list (1,2). All individuals underwent genetic testing for noncardiac reason and no documented incidence of CVD. According to the German data protection and gene diagnostic law, we reported the pathogenic variants in actionable genes listed by ACMG (1). Variants of unknown significance (VUS), whose involvement in disease at the current time was unclear, were not reported. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by local institutions (2019-091). Informed consent was taken from all the patients.
Gene list for screening of secondary findings
NGS data were analysed for variants in actionable genes of the ACMG SF v2.0 list (1,2) associated with CVD including EDS type IV, LDS, MFS, fTAAD, HCM, DCM, LVNC, ARVC, Fabry's disease, CPVT, LQTS and BrS. The list of the selected genes includes: ACTA2 (NM_001613.2), ACTC1 (NM_005159.4), COL3A1 (NM_000090.3), DSC2 (NM_004949.4), DSG2 (NM_001943.4), DSP (NM_004415.3), FBN1 (NM_000138.4), GLA (NM_000169.2), KCNH2 (NM_000238.3), KCNQ1 (NM_000218.2), LMNA (NM_005572.3), MYBPC3 (NM_000256.3), MYH11 (NM_002474.2), MYH7 (NM_000257.3), MYL2 (NM_000432.3), MYL3 (NM_000258.2), PKP2 (NM_004572.3, NG_009000.1), PRKAG2 (NM_016203.3), RYR2 (NM_001035.2), SCN5A (NM_198056.2), SMAD3 (NM_005902.3), TGFBR1 (NM_004612.3), TGFBR2 (NM_001024847.2), TMEM43 (NM_024334.2), TNNI3 (NM_000363.4), TNNT2 (NM_001001430.2), TPM1 (NM_001018005.1).
High throughput sequencing and bioinformatics pipeline
Next-generation sequencing (NGS) analysis of a custom capture kit (Agilent SureSelectXT) was carried out on an Illumina NextSeq 500 system (Illumina, San Diego, CA) as 150 bp paired-end sequencing runs using v2.0 SBS chemistry. Sequencing reads were aligned to the human reference genome (GRCh37/hg19) using BWA (v0.7. 13-r1126) with standard parameters. SNV, CNV and INDEL calling on the genes was conducted using the varvis software platform (varvis™, Limbus Technologies) with subsequent coverage and quality dependent filter steps.
Nomenclature, interpretation and classification of genetic variants
The nomenclature guidelines of the Human Genome Variation Society (HGVS) were used to annotate DNA sequence variants (31). The functional consequence of missense variants was interpreted with the amino acid (AA) substitution effect prediction methods SIFT (Sorting Invariant from Tolerated), PolyPhen-2, Mutation Taster and MAPP. Splice-sites were predicted with MES and SSF. Population databases were used to assess the allele frequencies of the variants: Database of all known Single Nucleotide Polymorphisms (dbSNP153, https://www.ncbi.nlm.nih.gov/snp/) and Genome Aggregation Database (gnomAD v2.2.1, http://gnomad.broadinstitute.org). GnomAD contains variants from whole-genome sequencing (WGS) of 71,702 samples from several large-scale projects of various disease-specific and population genetic studies.
The identified variants were classified according to the ACMG guidelines with the 5-tier classification system: class 5 (pathogenic), class 4 (likely pathogenic), class 3 (variants of unknown significance, VUS), class 2 (likely benign) and class 1 (benign) (32). Variants classified as likely pathogenic (class 4) or pathogenic (class 5) are named uniformly as pathogenic in the following study. Variant classification was compared by common databases such as ClinVar, LOVD and ARVC genetic variants database. ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) is a freely accessible, public archive of reports of the relationship among human variations and phenotypes, with supporting evidence (33). The ARVC database is a freely available collection of variants associated with ARVC and can be accessed via the link http://www.arvcdatabase.info/ (34). LOVD (Leiden Open (source) Variation Database is also a free, open source database (https://www.lovd.nl/).
Data availability statement
The variants in actionable ARVC genes are available at https://databases.lovd.nl/shared/variants/DSC2, https://databases.lovd.nl/shared/variants/DSG2, https://databases.lovd.nl/shared/variants/DSP, https://databases.lovd.nl/shared/variants/PKP2, https://databases.lovd.nl/shared/variants/TMEM43
Results
In 1% of 6,605 next-generation sequencing analyses pathogenic secondary findings associated with inherited heart or vascular disease were identified
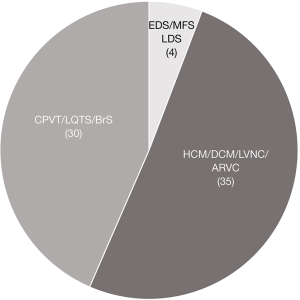
We analyzed NGS data of 6,605 individuals (unrelated to CVD) for pathogenic variants in one of the selected 27 actionable genes associated with CVD of the ACMG v2.0 list. 1% (69 of 6,605) of these individuals were found to harbor a pathogenic variant in one of the genes. We identified 30 pathogenic variants in genes causing arrhythmia syndromes (LQTS, BrS, CPVT), 22 pathogenic variants in genes associated with cardiomyopathies (HCM, DCM, LVNC), 13 pathogenic variants in genes causing ARVC, three pathogenic variants in COL3A1 associated with EDS type 4, one pathogenic variant in SMAD3 associated with LDS type 3. No pathogenic variant was present in FBN1, associated with MFS, none in GLA associated with Fabry’s disease and none in genes associated with fTAAD. Overall, 94% (65/69) of all pathogenic SF were linked to cardiomyopathies or arrhythmia syndromes and 6% (4/69) of all SF were detected in genes linked to vascular and connective tissue disease such vascular EDS and LDS type 3 (Figure 1).
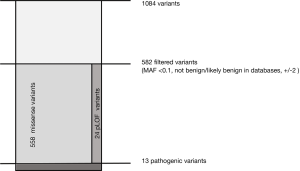
Gene-specific interpretation of secondary findings in PKP2, DSC2, DSG2, DSP and TMEM43
ARVC is the most common cause of life-threatening arrhythmias and SCD in young adults and athletes (12,35). The presence of a pathogenic variant is a major criterion to establish diagnosis (16). We therefore focused on the filtering procedure and interpretation of variants in the ARVC-associated genes: PKP2, DSC2, DSG2, DSP and TMEM43 (Figure 2). In a first filter step (MAF <0.01, variants in exons +/- 10bp) we identified a mean value of 1,084 variants in all five ARVC genes (Figure 2). Next, all variants that have been classified as benign or likely benign (class 1 and 2 according to ACMG) in common databases and all silent and non-coding variants (> +/- 2bp from exon) were filtered out. After this filtering step we ended up with 582 variants (class 3, 4 and 5) including 558 (96%) missense variants and 24 (4%) putative loss of function (pLoF) variants (stop-gain, start-loss, splice and frameshift variants) (Figure 2). From all rare variants identified, 13 pLoF variants were selected for return as pathogenic actionable SF but none of the missense variants could be classified as pathogenic and therefore were not returned to the patients. The decision was made based on the known pathogenicity mechanism of disease where haploinsufficiency is essential for disease manifestation, multiple lines of computational evidence (conservation, evolutionary, splicing impact, etc.) and the listing of variants as pathogenic by multiple submitters in different databases (ClinVar, ARVC, LOVD) (Figure 2).
Eight of the 24 pLoF variants have been previously reported to cause ARVC (known pathogenic, KP), 16 of the variants have not been described before. Five of the novel variants are expected to cause ARVC (EP) and eleven variants are of unknown significance (VUS) causing ARVC (Table S1). We classified the variants according to the specified ACMG/AMP recommendations for interpreting the loss of function PVS1 variant criterion (36) (Table S1).
We identified three pLoF in DSP that have not been described before. The variant [c.7745_7746del p.(Phe2582*)] is located in the last exon of DSP, 871 bp upstream of the stop codon, with other known pathogenic variants located downstream, assuming that nonsense mediated decay (NMD) is not predicted to occur. This variant probably results in a stable mRNA, which is not prone to NMD and directs the synthesis of a C-terminally truncated polypeptides, destroying the plectin repeat of DSP, a region critically for protein function which may result in ARVC. Therefore this variant is expected to be pathogenic (EP, class 4). The two other identified nonsense variants [c.8451C>A p.(Tyr2817*); c.8494G>T p.(Gly2832*)] are also located in the last exon of DSP, but downstream of the most 3′ truncating known pathogenic variant and downstream of the plectin region. Therefore both variants were classified as class 3 (VUS) and were not reported as actionable SF (Table S1).
In DSC2 we found four truncating variants: one start-loss [c.2T>A, (p.?)], two stop-gain [c.34G>T p.(Gly12*); c.1777G>T p.(Glu593*)] and one frameshift variant [c.2530_2531del p.(Leu844Aspfs*2)]. All variants have not been described in a database before. The two stop-gain variants are expected to be pathogenic (EP), because LoF of DSC2 is a known pathomechanism of ARVC. Pathogenic stop-gain variants are found in the databases up and downstream of the new identified variants. The start los variant c.2T>A, (p.?) and the frameshift variant c.2530_2531del:p.(Leu844Aspfs*2) could not be classified as pathogenic, because in both cases the strength level of PVS1 must be decreased to moderate (PVS1_moderate) (Table S1).
In DSG2, we identified three pathogenic truncating variants and one truncating variant with unknown significance. Two known pathogenic variants (KP), were reported in individuals affected with ARVC (37-39): one start-loss variant [c.3G>A, (p.?)] and one frameshift variant [c.3059_3062del p.(Glu1020Alafs*18)]. One novel expected pathogenic stop gain variant c.3025C>T p.(Gln1009*) is located in the last exon 332bp upstream of the stop codon, which has not been described in the literature before. In this case, pathogenic variants downstream of the new stop codon supports the biological relevance of the missing C terminal region. The stop-gain variant of unknown significance [c.3340C>T p.(Gln1114*)] is located at the end of the last exon only 11 bp upstream of the stop codon. No pathogenic stop variant has been published downstream of this variant, therefore the missing three C-terminal amino acids in the resulting protein are probably not critical for protein function. Therefore the variant could not classified as pathogenic (Table S1).
In PKP2 we identified two splice variants c.2146-1G>C and c.1378+1G>C, one stop-gain variant [c.1138G>T p.(Glu380*)] and two frameshift variants [c.1211dup p.(Val406Serfs*4); c.1664del p.(Phe555Serfs*8)]. All variants have been reported in individuals affected with ARVC in the literature. One variant with a predicted moderate change at the splice donor site 1 bp downstream, located in exon 1 could not be classified as pathogenic (Table S1).
In TMEM43, no pLoF variant and no missense variant was classified as pathogenic in our cohort. TMEM43 is a gene, were LoF is not a known mechanism of disease. No pathogenic missense variant could be identified in this gene.
Identification of the variant type in ARVC cases
To interpret gene-specific findings, we asked for the variant types identified in ARVC patients in DSC2, DSG2, DSP, PKP2 and TMEM43. Therefore we evaluated pathogenic variants listed in the ClinVar and the ARVC database (Figure 3). Overall, 700 variants (class 3, 4 and 5), including 63% missense and 37% pLoF variants were listed in the ARVC database. 29% of the missense variants and 89% of the pLoF variants were classified as pathogenic in all five ARVC genes (Figure 3A). 83% of the pLoF variants were classified as pathogenic in DSC2, 82% in DSG2, 80% in DSP and 97% in PKP2 (Figure 3A). In TMEM43 only two splice variants (one pathogenic and one VUS) and no truncating variant was listed in the ARVC database.
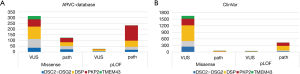
In ClinVar, 2178 variants (class 3, 4 and 5), including 78% missense and 22% pLoF variants were listed. 3% of the missense variants and 90% of the pLoF were classified as pathogenic in all five ARVC genes (Figure 3B). 74% of the pLoF variants listed in DSC2, 80% in DSG2, 96% in DSP and 96% in PKP2 (Figure 3B). In ClinVar, 14 pLoF variants were listed in TMEM43, none of these variants were classified as pathogenic for ARVC (one pLoF located in the last exon was found once in a patient with cardiomyopathy), also indicating that LoF is not a pathomechanism in TMEM43-associated ARVC. But the overall prevalence of variants in this gene is very low (Figure 3B).
Finally, we searched in the population database gnomAD, for all identified variants in the five ARVC genes. Overall, 8,050 variants were listed in gnomAD. 45% of the variants were documented as missense variants and 3% (2–5%) as pLoF variants, 52% of the variants were intronic variants (> +/-10bp from exon) and silent variants (Data not shown).
Discussion
Here, we determined the frequency of medically actionable SF in genes linked to CVD and highlight the challenge evaluating new variants in actionable ARVC genes in non affected patients, by analyzing sequencing data of 6,605 individuals who underwent genetic testing for noncardiac diagnostic request. Overall, 69 of 6,605 (1%) individuals carried pathogenic SF in one of the 27 actionable genes from the ACMG SF v2.0 list associated to CVD. Of them, 0,2% of 6,605 individuals carried a pathogenic variant in one of the five actionable ARVC genes (DSC2, DSG2, DSP, PKP2 and TMEM43). 9% (582) of 6,605 individuals carried a rare variant in one of the ARVC genes. From all rare variants identified, 96% (558/582) were missense variants and 4% (24/582) putative LoF (pLoF) variants.
Due to the fact that ARVC is associated with a high risk of SCD, decision to return an actionable genetic finding for ARVC has particular challenges. The SF Working Group recommends the return of known pathogenic (KP) and expected pathogenic (EP) variants in PKP2, DSP, DSC2 and DSG2. Importantly, for TMEM43 only KP variants are recommended to be returned as SF (6).
Since EP variants are of the type which is expected to cause the disorder, we evaluated the predominant variant type that is expected to cause ARVC. Therefore, we determined the frequency of the variant type in ARVC cases listed in ClinVar and ARVC database. In ClinVar, 90% of the pLoF variants and 3% of the missense variants were classified as pathogenic in the five ARVC genes. From all variants listed in the ARVC database, 89% of the pLoF variants and 28% missense variants were classified as pathogenic in all five ARVC genes. The data indicate that LoF is probably the predominant pathomechanism in ARVC and pLoF variants identified as SF should be considered for further evaluation, with the exception of TMEM43, were LoF variants have no supportive evidence for pathogenicity. Since one-third of all variants listed in ARVC database are missense variants, our data further highlight the need for a careful interpretation of seemingly class 3 (VUS) missense variants detected as SF in ARVC. Since our knowledge expands, a system needs to be in place for the review of genetic findings and VUS should be upgraded to disease-causing or downgraded to benign in particular with regard to treatable arrhythmic disorders with risk of SCD.
In line, with the data from ClinVar and ARVC database, a low frequency of pLoF variants was found in the population-based database gnomAD. Importantly, the constraint score shown in gnomAD (the ratio of the observed/expected number of LoF variants in a gene) do not per se indicate that ARVC genes are LoF intolerant, but this is in the expected range since ARVC penetrance is reduced.
In our study, we identified 24 (0.4%) pLoF variants in 6,605 individuals. From all 24 identified pLoF variants, eight were KP variants and five were EP variants. From all rare variants identified in TMEM43 in our cohort, neither missense nor pLoF variants, have been classified as pathogenic. Most of the pathogenic variants listed in TMEM43 were missense variants. In particular the missense variant, c.1073C>T p.(Ser358Leu) in TMEM43, has been genetically identified to cause ARVC type 5, a fully penetrant, lethal arrhythmic disorder. This founder mutation is located in a highly conserved transmembrane domain of TMEM43 and was first identified in families in Newfoundland by Merner et al. (40). These data indicate a critical assessment of variants in TMEM43 identified as SF. We suggest that only specific variants (KP) in TMEM43 should be listed as actionable findings in the recommended SF list.
Variants predicted to result in LoF have attracted interest because of their clinical impact and surprising prevalence in healthy individuals. pLoF variants are predicted to seriously disrupt the function of human protein coding genes and are frequently associated with ARVC. These variants include stop-gain, indel frameshift or essential splice-site disruption. Haploinsufficiency may arise from a LoF in the variant allele, such that it produces little or no gene product. Premature termination codon-introducing variants cause human genetic diseases, making NMD an important modulator of disease outcome. The NMD pathway is an mRNA surveillance system that typically degrades transcripts containing premature termination codons (PTCs) in order to prevent translation of possible dominant negative or aberrant transcripts, resulting in a predicted LoF variant. Activation of NMD depends on the position of the variant. Generally truncating variants within the last exon and 50 nt of the penultimate exon as well as in the first 200 nt of a transcript (if an additional in-frame ATG is present) may escape NMD and possibly yield a stable mRNA that directs the synthesis of truncated polypeptides (41) that may result in disease due to gain-of-function effects.
pLoF variants of different genes were curated by disease-specific working groups. The NIH-funded Clinical Genome Resource (ClinGen) established the Sequence Variant Interpretation (SVI) working group (https://www.clinicalgenome.org/working-groups/sequence-variant-interpretation/) to refine and evolve the ACMG rules for accurate and consistent clinical application. The ClinGen dosage sensitivity curation process collects evidence supporting the haploinsufficiency and triplosensitivity of genes and genomic regions. The ClinGen haploinsufficiency scores (score 1–3) for an individual gene may be used to guide the clinical interpretation of deletions involving a particular gene (ClinGen Dosage Sensitivity Map).
Since pLoF variants have a high probability of disease association (42) a study by Haggerty and co-workers evaluated a genotype-phenotype association in a large unselected cohort (30,716 individuals) with identified pLoF variants by electronic health record (EHR) (43). The study showed that eighteen subjects had pLoF variants and none of them had an EHR diagnosis of ARVC. Of 14 patients with an ECG, one had a minor diagnostic criterion of ARVC (Task Force criteria), the remaining were normal. Those data indicate that, in unselected individuals with an actionable SF, genetic penetrance may be lower than expected (40–60%) from familial studies (44-46).
Haedrick and colleagues (30) evaluated SF in ARVC genes in children who underwent WES testing for noncardiac disease. The study reported that rare variants associated with ARVC occurred in 14% of the WES cohort. The vast majority of variants within the WES cohort were missense variants (83.7%). A higher frequency of high impact variants were found in ARVC cases. Review of clinical data available on WES referrals demonstrated none with evidence of ARVC among variant-positive individuals. Data further highlight the need for a careful interpretation of missense variants detected as SF in ARVC. Nevertheless, the study could not exclude that some of the retrospectively reviewed children my yet develop ARVC. Since ARVC exhibits an age-dependent penetrance, with symptoms and diagnostic criteria developing with time (46), a lifelong follow-up is needed. Quarta et al. (47) reported that the cumulative prevalence of ARVC is essentially flat after 60 years of age.
Interpretation of SF in ARVC-related genes becomes even more challenging since also digenic, homozygous, and compound heterozygous inheritance has been discussed (42,48,49) which might be associated with a more severe disease course and with a higher incidence and earlier onset of sustained arrhythmia and an increased risk of SCD (44,49).
In our study none of the individuals carried more than one pathogenic variant associated with ARVC. While compound or digenic heterozygosity clearly plays an important role in explaining the incomplete penetrance and variable expressivity of ARVC, we believe that the type and location of the variant might be similarly important. Moreover, the full phenotypic spectrum of variants in desmosomal genes has not completely understood. Pathogenic missense variants in PKP2 have been published in patients with BrS (50) and CPVT (51) variants in DSP have also been observed in patients with DCM.
One of the diagnostic challenges with ARVC is the lack of sensitive techniques to reflect a subclinical or concealed phase of ARVC. New diagnostic innovations are needed for early detection in genotype-positive/phenotype-negative cases. Perrin et al. (52) determined if exercise treadmill testing (ETT) could expose a latent substrate of ARVC in asymptomatic carriers. Exercise-induced abnormalities during ETT were initially compared in 60 subjects (30 asymptomatic carriers of pathogenic variants in ARVC genes and 30 healthy controls). Results show that exercise testing exposes a latent electrical substrate (depolarization abnormalities during ETT) in asymptomatic carriers that is shared by ARVC patients with histories of ventricular arrhythmia (VA). This data indicates that, ETT may be useful in guiding treatment decisions, exercise prescription, and prioritizing medical surveillance in asymptomatic carriers.
After diagnosis, therapeutic options for ARVC patients consist of pharmacological treatment (e.g., antiarrhythmics or beta-blockers), implantable cardioverter defibrillator (ICD) placement, catheter ablation and lifestyle modifications. Corrado et al. published a comprehensive description of guidelines and recommendations for treatment (53). In ARVC there is the unique opportunity to reduce the likelihood of developing ARVC through lifestyle modification. Exercise has a well-established role in the pathogenesis of ARVC, and recognition of a desmosomal gene variant can help to determine optimal recommendations (54). Recent studies have shown a link between frequent physical activity and VA, heart failure and transplant, lifestyle modification through avoidance of endurance exercise is recommended (27,54,55).
There is a wide range of opinions about SF in clinical sequencing and how they should be managed. On one side patients have the right to be informed about possible risks. On the other side there is insufficient evidence about the penetrance of most pathogenic variants in the general population and return of SF creates the psychological burden of being a “patient in waiting” (56).
Whenever clinical sequencing is ordered, the ordering clinician should discuss with the patient the possibility of identifying SF. The informed consent process for clinical sequencing should follow the forthcoming guideline from the ACMG. In particular, the return of SF to parents of minor children who undergo clinical sequencing presents difficult issues. The Working Group recommended that recommendations for seeking and reporting SF not be limited by the age of the person being sequenced (6). To withhold SF is to state that the child’s right not to know supersedes the parent’s opportunity to discover a life-threatening risk factor (6). Patients have the right to decline clinical sequencing if they judge the risks of possible discovery of SF to outweigh the benefits of testing. It is the responsibility of the ordering team to provide comprehensive pre- and post-test counseling to the patient and clinician should also provide medical follow-up as described in the prior ACMG policy statement on Clinical Application of Genomic Sequencing (57). Given the complexity of genomic information, the clinical geneticist should be consulted at the appropriate time that may include ordering interpreting, and communicating genomic testing (28,58).
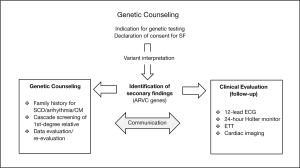
We would recommend collaboration and multidisciplinary team (MDT) meetings with a collective statement before reporting a SF. If decided to return a SF to patient, an effective management by a MDT approach, in cooperating expertise in inherited heart disease should be offered (Figure 4). The clinical specialist decides which medical follow-up (e.g., 12-lead ECG, 24-hour Holter monitor, ETT, cardiac imaging) should be provided. Since research on genetics is rapidly advancing, the genetic specialist should be consulted on a regular interval, to critically assess SF in a context of clinical manifestation.
Conclusions
Our data highlight the importance of a careful variant interpretation in actionable genes recommended for return as SF. The vast majority of true pathogenic SF in ARVC genes is rare. Each variant must undergo rigorous clinical and laboratory evaluation before it can be described as pathogenic and returned to patients. Variants of uncertain clinical significance should not be returned as SF to patient. Gene-specific interpretation of SF in ARVC-related genes by expert disease-specific knowledge has a great impact to provide diagnosis and subsequently prevention of SCD.
Acknowledgments
We thank Kristina Lenhard for technical assistance.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Yskert von Kodolitsch, Harald Kaemmerer, Koichiro Niwa) for the series “Current Management Aspects in Adult Congenital Heart Disease (ACHD): Part III” published in Cardiovascular Diagnosis and Therapy. The article has undergone external peer review.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/cdt-20-585
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-585). The series “Current Management Aspects in Adult Congenital Heart Disease (ACHD): Part III” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by local institutions (2019-091). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19:249-55. [Crossref] [PubMed]
- Biesecker LG. ACMG secondary findings 2.0. Genet Med 2017;19:604. [Crossref] [PubMed]
- Wolf SM, Crock BN, Van Ness B, et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med 2012;14:361-84. [Crossref] [PubMed]
- Fullerton SM, Wolf WA, Brothers KB, et al. Return of individual research results from genome-wide association studies: experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genet Med 2012;14:424-31. [Crossref] [PubMed]
- Angrist M. You never call, you never write: why return of 'omic' results to research participants is both a good idea and a moral imperative. Per Med 2011;8:651-7. [Crossref] [PubMed]
- Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565-74. [Crossref] [PubMed]
- Basso C, Bauce B, Corrado D, et al. Pathophysiology of arrhythmogenic cardiomyopathy. Nat Rev Cardiol 2011;9:223-33. [Crossref] [PubMed]
- Corrado D, Link MS, Calkins H. Arrhythmogenic Right Ventricular Cardiomyopathy. N Engl J Med 2017;376:1489-90. [Crossref] [PubMed]
- Basso C, Corrado D, Marcus FI, et al. Arrhythmogenic right ventricular cardiomyopathy. Lancet 2009;373:1289-300. [Crossref] [PubMed]
- Olfson E, Cottrell CE, Davidson NO, et al. Identification of Medically Actionable Secondary Findings in the 1000 Genomes. PLoS One 2015;10:e0135193 [Crossref] [PubMed]
- Sen-Chowdhry S, Morgan RD, Chambers JC, et al. Arrhythmogenic cardiomyopathy: etiology, diagnosis, and treatment. Annu Rev Med 2010;61:233-53. [Crossref] [PubMed]
- Corrado D, Basso C, Schiavon M, et al. Screening for hypertrophic cardiomyopathy in young athletes. N Engl J Med 1998;339:364-9. [Crossref] [PubMed]
- Dalal D, Nasir K, Bomma C, et al. Arrhythmogenic right ventricular dysplasia: a United States experience. Circulation 2005;112:3823-32. [Crossref] [PubMed]
- Romero J, Mejia-Lopez E, Manrique C, et al. Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC/D): A Systematic Literature Review. Clin Med Insights Cardiol 2013;7:97-114. [Crossref] [PubMed]
- Bagnall RD, Weintraub RG, Ingles J, et al. A Prospective Study of Sudden Cardiac Death among Children and Young Adults. N Engl J Med 2016;374:2441-52. [Crossref] [PubMed]
- Marcus FI, McKenna WJ, Sherrill D, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J 2010;31:806-14. [Crossref] [PubMed]
- Sen-Chowdhry S, Syrris P, McKenna WJ. Role of genetic analysis in the management of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol 2007;50:1813-21. [Crossref] [PubMed]
- Gerull B, Heuser A, Wichter T, et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet 2004;36:1162-4. [Crossref] [PubMed]
- Syrris P, Ward D, Evans A, et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in the desmosomal gene desmocollin-2. Am J Hum Genet 2006;79:978-84. [Crossref] [PubMed]
- Awad MM, Dalal D, Cho E, et al. DSG2 mutations contribute to arrhythmogenic right ventricular dysplasia/cardiomyopathy. Am J Hum Genet 2006;79:136-42. [Crossref] [PubMed]
- Rampazzo A, Nava A, Malacrida S, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet 2002;71:1200-6. [Crossref] [PubMed]
- McKoy G, Protonotarios N, Crosby A, et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 2000;355:2119-24. [Crossref] [PubMed]
- Lorenzon A, Pilichou K, Rigato I, et al. Homozygous Desmocollin-2 Mutations and Arrhythmogenic Cardiomyopathy. Am J Cardiol 2015;116:1245-51. [Crossref] [PubMed]
- Qadri S, Anttonen O, Viikila J, et al. Case reports of two pedigrees with recessive arrhythmogenic right ventricular cardiomyopathy associated with homozygous Thr335Ala variant in DSG2. BMC Med Genet 2017;18:86. [Crossref] [PubMed]
- Hodgkinson KA, Connors SP, Merner N, et al. The natural history of a genetic subtype of arrhythmogenic right ventricular cardiomyopathy caused by a p.S358L mutation in TMEM43. Clin Genet 2013;83:321-31. [Crossref] [PubMed]
- van der Zwaag PA, van Rijsingen IA, Asimaki A, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail 2012;14:1199-207. [Crossref] [PubMed]
- Haggerty CM, Murray B, Tichnell C, et al. Managing Secondary Genomic Findings Associated With Arrhythmogenic Right Ventricular Cardiomyopathy: Case Studies and Proposal for Clinical Surveillance. Circ Genom Precis Med 2018;11:e002237 [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Hall CL, Sutanto H, Dalageorgou C, et al. Frequency of genetic variants associated with arrhythmogenic right ventricular cardiomyopathy in the genome aggregation database. Eur J Hum Genet 2018;26:1312-8. [Crossref] [PubMed]
- Headrick AT, Rosenfeld JA, Yang Y, et al. Incidentally identified genetic variants in arrhythmogenic right ventricular cardiomyopathy-associated genes among children undergoing exome sequencing reflect healthy population variation. Mol Genet Genomic Med 2019;7:e593 [Crossref] [PubMed]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet 2001;109:121-4. [Crossref] [PubMed]
- Matthijs G, Souche E, Alders M, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet 2016;24:2-5. [Crossref] [PubMed]
- Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44:D862-8. [Crossref] [PubMed]
- Lazzarini E, Jongbloed JD, Pilichou K, et al. The ARVD/C genetic variants database: 2014 update. Hum Mutat 2015;36:403-10. [Crossref] [PubMed]
- Corrado D, Fontaine G, Marcus FI, et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy: need for an international registry. Study Group on Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy of the Working Groups on Myocardial and Pericardial Disease and Arrhythmias of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the World Heart Federation. Circulation 2000;101:E101-6. [PubMed]
- Abou Tayoun AN, Pesaran T, DiStefano MT, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat 2018;39:1517-24. [Crossref] [PubMed]
- Christensen AH, Benn M, Bundgaard H, et al. Wide spectrum of desmosomal mutations in Danish patients with arrhythmogenic right ventricular cardiomyopathy. J Med Genet 2010;47:736-44. [Crossref] [PubMed]
- Lahtinen AM, Lehtonen E, Marjamaa A, et al. Population-prevalent desmosomal mutations predisposing to arrhythmogenic right ventricular cardiomyopathy. Heart Rhythm 2011;8:1214-21. [Crossref] [PubMed]
- Rasmussen TB, Palmfeldt J, Nissen PH, et al. Mutated desmoglein-2 proteins are incorporated into desmosomes and exhibit dominant-negative effects in arrhythmogenic right ventricular cardiomyopathy. Hum Mutat 2013;34:697-705. [Crossref] [PubMed]
- Merner ND, Hodgkinson KA, Haywood AF, et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet 2008;82:809-21. [Crossref] [PubMed]
- Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 2016;48:1112-8. [Crossref] [PubMed]
- Kapplinger JD, Landstrom AP, Salisbury BA, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol 2011;57:2317-27. [Crossref] [PubMed]
- Haggerty CM, James CA, Calkins H, et al. Electronic health record phenotype in subjects with genetic variants associated with arrhythmogenic right ventricular cardiomyopathy: a study of 30,716 subjects with exome sequencing. Genet Med 2017;19:1245-52. [Crossref] [PubMed]
- Groeneweg JA, Bhonsale A, James CA, et al. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circ Cardiovasc Genet 2015;8:437-46. [Crossref] [PubMed]
- te Riele AS, Bhonsale A, James CA, et al. Incremental value of cardiac magnetic resonance imaging in arrhythmic risk stratification of arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J Am Coll Cardiol 2013;62:1761-9. [Crossref] [PubMed]
- te Riele AS, James CA, Groeneweg JA, et al. Approach to family screening in arrhythmogenic right ventricular dysplasia/cardiomyopathy. Eur Heart J 2016;37:755-63. [Crossref] [PubMed]
- Quarta G, Muir A, Pantazis A, et al. Familial evaluation in arrhythmogenic right ventricular cardiomyopathy: impact of genetics and revised task force criteria. Circulation 2011;123:2701-9. [Crossref] [PubMed]
- Xu T, Yang Z, Vatta M, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol 2010;55:587-97. [Crossref] [PubMed]
- Bhonsale A, Groeneweg JA, James CA, et al. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. Eur Heart J 2015;36:847-55. [Crossref] [PubMed]
- Cerrone M, Lin X, Zhang M, et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation 2014;129:1092-103. [Crossref] [PubMed]
- Tester DJ, Ackerman JP, Giudicessi JR, et al. Plakophilin-2 Truncation Variants in Patients Clinically Diagnosed With Catecholaminergic Polymorphic Ventricular Tachycardia and Decedents With Exercise-Associated Autopsy Negative Sudden Unexplained Death in the Young. JACC Clin Electrophysiol 2019;5:120-7. [Crossref] [PubMed]
- Perrin MJ, Angaran P, Laksman Z, et al. Exercise testing in asymptomatic gene carriers exposes a latent electrical substrate of arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol 2013;62:1772-9. [Crossref] [PubMed]
- Corrado D, Wichter T, Link MS, et al. Treatment of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia: An International Task Force Consensus Statement. Circulation 2015;132:441-53. [Crossref] [PubMed]
- James CA, Bhonsale A, Tichnell C, et al. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J Am Coll Cardiol 2013;62:1290-7. [Crossref] [PubMed]
- Saberniak J, Hasselberg NE, Borgquist R, et al. Vigorous physical activity impairs myocardial function in patients with arrhythmogenic right ventricular cardiomyopathy and in mutation positive family members. Eur J Heart Fail 2014;16:1337-44. [Crossref] [PubMed]
- Kwon JM, Steiner RD. "I'm fine; I'm just waiting for my disease": the new and growing class of presymptomatic patients. Neurology 2011;77:522-3. [Crossref] [PubMed]
- Directors ABo. Points to consider in the clinical application of genomic sequencing. Genet Med 2012;14:759-61. [Crossref] [PubMed]
- Towbin JA, McKenna WJ, Abrams DJ, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm 2019;16:e301-72. [Crossref] [PubMed]


