A machine learning-based approach for the prediction of periprocedural myocardial infarction by using routine data
Introduction
Cardiovascular diseases (CVDs) are one of the leading causes of mortality and morbidity all over the world, about 17.3 million people died in 2013 due to CVD, higher than that in 1990 (1). In the last few decades, percutaneous coronary intervention (PCI) is becoming the most popular treatment for coronary artery diseases (CADs) (2). Although technical advances in PCI process, there is still a high incident rate of periprocedural myocardial infarction (PMI) about 5–30% in the different studies (3,4). Moreover, clinical practitioners have tried their best to find the relationship between preprocedural indicators and the incidence of PMI. Unfortunately, it isn’t established that the change of a single factor or some specific factors could predict the presence of PMI.
Hopefully, machine learning (ML) provides a better way to explore the relationship between large scale baseline data and the incidence of PMI. ML can be referred as a general-purpose system with a capability of reasoning and thinking skills mimicking a human being’s brain (5), and this approach relies on computers to exploit all complex and non-linear interactions across all the attributes to build the best model for the prediction of observed outcomes (6). In practice, ML has shown its power in the prediction of the risk of CVD in 10-year time period in the UK (7), prognosis in the heart failure population in the USA (8), and the length of stay in Arabian countries (9). Many investigators believe ML methods could build a more personalized and precise model based on the big dataset.
To date, these has been no large-scale investigation applying ML methods for prediction of PMI in CAD population by using preprocedural clinical data. The aim of this research is to evaluate whether ML can develop a robust prediction model for PMI. And we also plan to determine which class of ML algorithm has the highest predictive potential. We present the following study in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/cdt-20-551).
Methods
Study setting and population
The dataset of this research was based on the inpatients who were admitted in department of cardiology of Sir Run Run Shaw hospital (Hangzhou, Zhejiang, China) from December 2007 to 2019 April. Since 2007, our department started to build an electronic medical system to record and evaluate the medical information for patients. Up to 2014, the electronic medical records system has been built for every department in our hospital to document outpatients and inpatients medical documentations, including demographic details, history of medical condition, bio-chemistry results, imaging impressions, primary diagnosis, prescription of drugs, records of interventions and surgeries, referrals of specialists, and following-up results. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Institutional Ethics Research Committee of Sir Run Run Shaw Hospital approved the study (approval ID: 20200224-33), and all patients provided written informed consent.
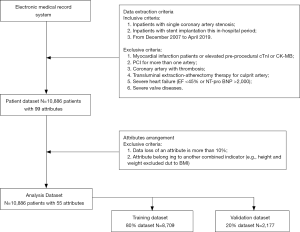
Data extraction and data preprocessing (Figure 1)
A total of 10,886 cardiac inpatients with 99 attributes were extracted from the electronic medical record system from December 2007 to 2019 April. The enrollment criteria as follows: (I) inpatients with single coronary artery stenosis (left main artery, left anterior descending artery, left circumflex artery, or right coronary artery); (II) inpatients with stent implantation this in-hospital period. The excluded criteria as follows: myocardial infarction patients or elevated pre-procedural cardiac troponin I (cTnI) or creatine kinase-MB fraction (CK-MB), PCI for more than one artery, coronary artery with thrombosis, transluminal extraction-atherectomy therapy for culprit artery, severe heart failure (EF <45% or NT-pro BNP >2,000), severe valve diseases.
About the attributes, these medical conditions related attributes were collected by experienced physicians and some laboratory results were recorded by trained technicians with standard automated machines. Also, all the results of each patient were collected in the last 24 hours before the procedure. And the cTnI and CK-MB levels were evaluated every 8 hours after the PCI, and a 24–48 hours dynamic monitoring would be acted after the procedure if necessary. However, if the data loss of an attribute was more than 10%, this attribute would be excluded. Moreover, if an attribute was belonged to another combined indicator [e.g., height and weight excluded due to body mass index (BMI)].
About the missing values, two methods were acted to do the missing data imputation, respectively (10). On the one side, the missing data would be excluded from the dataset, the remain data were used to build ML models. On the other side, mean imputation, another common approach to dealing with missing values in the ML was chose to modify the dataset.
Primary end point and definitions
The primary end point was PMI, according to the definitions of the Society for Cardiovascular Angiography and Interventions (SCAI) (11) and the Universal Definition of Myocardial Infarction (12), the Cut-Off value of cTnI for PMI was >3-fold or 5-fold upper reference limit (URL) after procedure in 48 hours. In our study, PMI3 represented the end point for cTnI >3-fold URL; PMI 5 represented the end point for cTnI >5-fold URL.
About the attributes, the top 20 attributes were selected based on the rank determined by the information gain method for the total 55 pre-procedural variables. After choosing the final 20 attributes, ML algorithms were acted to build models to predict the PMI patients.
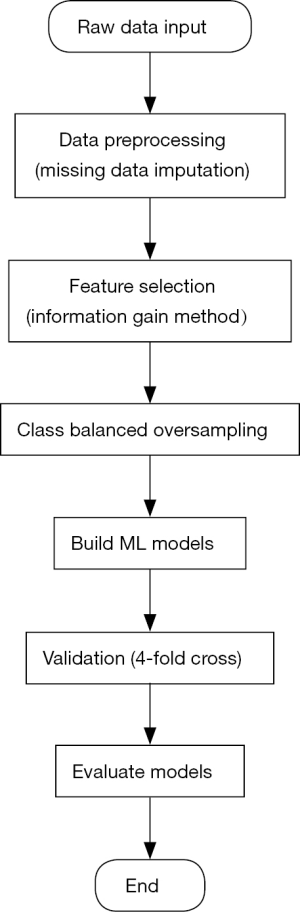
Feature selection and class balanced oversampling (Figure 2)
Feature selection was achieved by a method named as information gain. This method was acted in our study to rank the attributes of the dataset. Information gain measured how much information an attribute gave researchers about the outcome to be predicted. If an attribute was heterogenous in different predicted groups, it means that this attribute might be a powerful predictor in the model building process (13,14). And the value of an attribute’s information gain was defined as:
[1]
Where H(S) is the entropy of the dataset and H(Si) is the entropy of the i subset generated by partitioning dataset S based on a specific attribute X.
Class balanced oversampling method was another approach to balance the imbalanced dataset. Often real-word datasets were always composed of a large proportion of normal examples with a small proportion of abnormal examples (15). In our study, the population of PMI patients was much smaller than the that of negative patients, in order to have a discriminable and powerful prediction model, we used 1:1 (normal:abnormal) population to build models.
ML classification techniques
Generally, the performance of ML models can vary from different datasets, which had different variables and observed outcomes. Under this situation, four different ML algorithms [Support Vector Machine (SVM) (16), Logistic Regression (LR) (17), Random Forest (RF) (18), Artificial Neural Networks (ANN) (19)] are acted to build models in our study, moreover, the performance of each model would be determined by the area under curve (AUC) of ROC. All the algorithms were programmed and run under the Python 3.x software environment.
SVM is a classifier to aim to output an optimal hyperplane in the N-dimensional space (N is the number same as the number of attributes) to do the outcome prediction. LR is a statistical ML algorithm that classifies the data by considering outcome variables on strict final point and makes a logarithmic line that distinguishes between them. RF is an ensemble learning method for classification, regression and other tasks that operates by constructing a multitude of decision trees at training time and at testing to output the class that is the mode of the classes (classification). ANN tries to mimic the human brain in order to model complicated task with many interconnected nodes just like neurons in the brain. About the model evaluation and validation, 4-fold cross validation method was acted to check the models’ validation.
Statistical analysis
All data were collected by Statistical Package for the Social Science (SPSS) for macOS, version 23 (SPSS Inc., Chicago, IL, USA). Demographical characteristics of the study population were analyzed by SPSS. Categorical data were using the percentages to record, and assessed with chi-square test or Fisher’s exact test. Whereas continuous data were using the means ± standard deviations to record, and assessed with Student’s t-test. A P value less than 0.05 was considered to be statistically significant.
Results
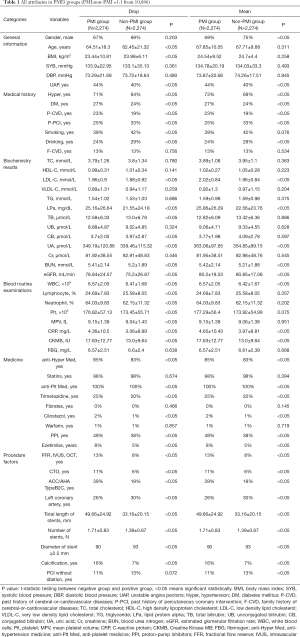
A total of (10,886) patients who were admitted in our hospital. PMI3 and PMI5 results were analyzed respectively. The incidence of PMI3 and PMI5 was 20.9% and 13.7%. Drop imputation and Mean imputation were individually applied in the dataset to build ML models. About PMI3, all attributes in the different groups were demonstrated on the Table 1, as well as PMI5 in the Table 2. All attributes were divided into six different categories (general information, medical history, biochemistry results, blood routine examinations, medicine, and procedure factors).
Full table

Full table
As an outcome of the feature selection process, information gain method was acted to rank the attributes’ importance to discriminate the non-PMI and PMI patients. Figure 3 showed all the results in different imputation datasets. The top 5 attributes in different groups all included eGFR, VLDL-C, BMI, and LPa.
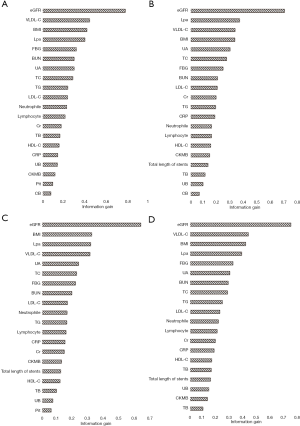
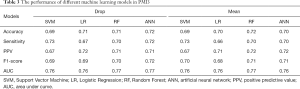
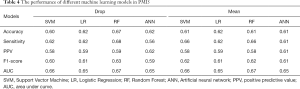
Table 3 summarized the performance of the different ML algorithms, and Table 4 showed the performance for PMI5. Generally, the ML algorithms’ performance in PMI3 was better than PMI5. Specifically, in PMI3 Drop group, ANN (accuracy: 0.72; AUC: 0.77) showed the best power to predict the presence of PMI; In PMI3 Mean Group, RF (accuracy: 0.72; AUC: 0.77) showed the best power; In PMI5 Drop group, RF (accuracy: 0.67; AUC: 0.67) showed the best power; In PMI5 Mean group, RF (accuracy: 0.61; AUC: 0.67) showed the best power.
Full table
Full table
Discussion
This study aimed to develop a ML-based model to predict the presence of PMI in CAD patients. A total of 10,886 patients were recruited in our study, six different categories data (general information, medical history, biochemistry results, blood routine examinations, medicine, and procedure factors) were used to consist of the dataset, four different algorithms (SVM, LR, RF, and ANN) were acted to build models to predict the PMI. ANN and RF showed the most accurate performance in PMI3 and PMI5.
Firstly, eGFR, VLDL-C, BMI, and LPa were four top indicators based on the analysis of information gain. In the previous studies, periprocedural TnT levels of eGFR <60 mL/min/1.73 m2 patients who received PCI were three times higher than the normal limit (20). About VLDL-C and LPa, remnant cholesterol could be valuable and independent predictor for PMI in diabetic patients (21), and non-HDL-C showed its power for the diagnosis of myocardial injury post procedurally (22). BMI was a combined index to assess people’s the degree of obesity, and an article reported that overweight patients had a better prognosis after primary angioplasty compared with other BMI groups. Information gain was a powerful method to explore the relationship between attributes and outcomes, not only just an important step to preprocess the data, but also may be used to assess the degree of relationship.
Secondly, different missing data imputation would lead to different results. In our study, two different methods for missing data imputation were acted to preprocess the data set, Drop the missing data and Median substitution. It is very hard to say which treatment for the missing data was better, moreover, our results also didn’t show the evidence for which method was better. Besides two ways in our study, other methods like k-nearest neighbors imputation and frequent values imputation, were also recommended in some articles (23,24). What’s more, multiple imputation was another way to replace missing data (25) in order to get a better dataset in some degree.
Thirdly, ML ANN and RF algorithms had a better power for prediction of PMI than ML SVM and LR algorithms in this research. In particular, there was no one-size-fits-all ML model. Literally, although RF and ANN had a better performance in this study, we cannot conclude that these two models would be better in other datasets. For example, in a Korean Study, deep neural networks were the best ML to predict the risk of CVD (26). And RF algorithm also showed its better power for prediction of in-hospital length (27). ANN showed its better performance in determining the risk of CVD in the UK population (7). Moreover, some articles reported that ML models were better than the traditional risk systems to predict the special outcomes, such as mortality of heart failure (8), acute coronary syndrome requiring revascularization (28), and risk of CVD mentioned before.
Fourthly, the performance of four ML models to predict PMI3 was better than that to predict PMI5. The difference between PMI3 dataset and PMI5 dataset was the data scale. Although we had 10,886 patients in our original dataset, about 80% of data were belonging to the negative group, after the oversampling step, ML algorithms cannot build a great model based on a small-scale dataset. In other words, datasets were the fundamental essential for a better prediction rather than the methods, including ML algorithms.
Limitations
Generally, there were several limitations of this current research. Firstly, as mentioned before, the dataset was from one-single health organization instead of several different centers. Secondly, it was acknowledged that the “black box” nature of ML models could be impossible for the interpretation of ML models. Thirdly, if the data loss of an attribute reached 10%, the attribute was removed from the dataset. This preprocess step would cause some biases before we knew the specific variable was significant for the prediction or not.
Conclusions
ML methods provide accurate prediction of PMI in CAD patients, and could be used as a precise model in the preventive treatment of PMI.
Acknowledgments
Xue Lv has given us a help with language editing.
Funding: This work was supported by the National Natural Science Foundation of China (81500212 and 81800212) and Zhejiang Natural Science Foundation (LY18H020007 and LQ17H020002).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/cdt-20-551
Data Sharing Statement: Available at http://dx.doi.org/10.21037/cdt-20-551
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-551). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Institutional Ethics Research Committee of Sir Run Run Shaw Hospital approved the study (approval ID: 20200224-33), and all patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117-71. [Crossref] [PubMed]
- Benjamin EJ, Virani SS, Callaway CW, et al. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association Circulation 2018;137:e67-e492. [published correction appears in Circulation. 2018 Mar 20;137(12):e493]. [Crossref] [PubMed]
- Herrmann J. Peri-procedural myocardial injury: 2005 Update. Eur Heart J 2005;26:2493-519. [Crossref] [PubMed]
- Testa L, Van Gaal WJ, Biondi Zoccai GGL, et al. Myocardial infarction after percutaneous coronary intervention: A meta-analysis of troponin elevation applying the new universal definition. QJM 2009;102:369-78. [Crossref] [PubMed]
- Seetharam K, Shrestha S, Sengupta PP. Artificial Intelligence in Cardiovascular Medicine. Curr Treat Options Cardiovasc Med 2019;21:25. [Crossref] [PubMed]
- Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: A methodology review. J Biomed Inform 2002;35:352-9. [Crossref] [PubMed]
- Weng SF, Reps J, Kai J, et al. Can Machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One 2017;12:e0174944. [Crossref] [PubMed]
- Panahiazar M, Taslimitehrani V, Pereira N, et al. Using EHRs and Machine Learning for Heart Failure Survival Analysis. Stud Health Technol Inform 2015;216:40-4. [PubMed]
- Daghistani TA, Elshawi R, Sakr S, et al. Predictors of in-hospital length of stay among cardiac patients: A machine learning approach. Int J Cardiol 2019;288:140-7. [Crossref] [PubMed]
- Batista GEAPA, Monard MC. An analysis of four missing data treatment methods for supervised learning. Appl Artif Intell 2003;17:519-33. [Crossref]
- Moussa ID, Klein LW, Shah B, et al. Consideration of a New Definition of Clinically Relevant Myocardial Infarction After Coronary Revascularization. J Am Coll Cardiol 2013;62:1563-70. [Crossref] [PubMed]
- Thygesen K, Alpert JS, Jaffe AS, et al. Fourth universal definition of myocardial infarction (2018). Russ J Cardiol 2019;24:107-38. [Crossref] [PubMed]
- Lee C, Lee GG. Information gain and divergence-based feature selection for machine learning-based text categorization. Inf Process Manag 2006;42:155-65. [Crossref]
- Kent J. Information Gain and a General Measure of Correlation. Biometrika 1983;70:163-73. [Crossref]
- Mordant N, Delour J, Léveque E, Arnéodo A, et al. Long time correlations in lagrangian dynamics: a key to intermittency in turbulence. Phys Rev Lett 2002;89:254502. [Crossref] [PubMed]
- Huang X, Maier A, Hornegger J, et al. Indefinite kernels in least squares support vector machines and principal component analysis. Appl Comput Harmon Anal 2017;43:162-72. [Crossref]
- Kleinbaum DG, Klein M. Logistic Regression A Self-Learning Text Second Edition., 2002.
- Classification and regression by randomForest. R News 2:18-22. Forest 2001;23(3).
- Yao X, Member S. Evolving Artificial Neural Networks. Proceedings of the IEEE 1999;87:1423-47. [Crossref]
- Kumagai S, Ishii H, Amano T, et al. Impact of chronic kidney disease on the incidence of peri-procedural myocardial injury in patients undergoing elective stent implantation. Nephrol Dial Transplant 2012;27:1059-63. [Crossref] [PubMed]
- Zeng RX, Li S, Zhang MZ, et al. Remnant cholesterol predicts periprocedural myocardial injury following percutaneous coronary intervention in poorly-controlled type 2 diabetes. J Cardiol 2017;70:113-20. [Crossref] [PubMed]
- Maadani M, Abdi S, Parchami-Ghazaee S, et al. Relationship between pre-procedural serum lipid profile and post-procedural myocardial injury in patients undergoing elective percutaneous coronary intervention. Res Cardiovasc Med 2013;2:169. [Crossref] [PubMed]
- Courrieu P, Rey A. Missing data imputation Chapter 25. Behav Res Methods 2011;43:310-30. [Crossref] [PubMed]
- Planned M, Protocols M, Design TPM, et al. Statistical Issues What Happens When Data 2015;I:760-97.
- Faris PD, Ghali WA, Brant R, et al. Multiple imputation versus data enhancement for dealing with missing data in observational health care outcome analyses. J Clin Epidemiol 2002;55:184-91. [Crossref] [PubMed]
- Kim J, Kang U, Lee Y. Statistics and deep belief network-based cardiovascular risk prediction. Healthc Inform Res 2017;23:169-75. [Crossref] [PubMed]
- Timóteo AT, Ramos R, Toste A, et al. Impact of body mass index in the results after primary angioplasty in patients with ST segment elevation acute myocardial infarction. Acute Card Care 2011;13:123-8. [Crossref] [PubMed]
- Noh YK, Park JY, Choi BG, et al. A Machine learning-based approach for the prediction of acute coronary syndrome requiring revascularization. J Med Syst 2019;43:253. [Crossref] [PubMed]