
MALAT1 gene rs600231 polymorphism positively associated with acute coronary syndrome in Chinese population: a case-control study
Introduction
Acute coronary syndrome (ACS) is the leading cause of cardiac mortality worldwide (1) and is characterized by myocardial ischaemia or necrosis, with an instantaneous decrease in coronary blood flow. Hence, it is critical that early detection and effective interventions reduce mortality and improve the prognosis of ACS (2). The current conventional myocardial injury biomarkers for ACS are creatine kinase MB (CK-MB) and cardiac troponin T (cTnT), which serve as the ‘gold standard’. However, these markers have limitations in terms of clinical sensitivity and specificity. For example, cTnT could be increased among individuals with other cardiovascular diseases (CVDs), such as severe heart failure, sepsis, or chronic renal insufficiency (3). The level of CK-MB also be increased when the skeletal muscle is damaged (4). In addition, circulating cTnT and CK-MB reach detectable levels after 4 hours and remain detectable for several days (5).Therefore, the identification of novel biomarkers is necessary for predicting ACS and improving clinical practice.
With the rapid development of high-throughput platforms, it has been shown that only 2% of genomic transcripts are in charge of coding proteins. Remarkably, a large portion of RNAs are grouped as long noncoding RNAs (lncRNAs). In recent years, lncRNAs >200 nucleotides in length have become valuable in research, and expanding evidence has indicated their biological function in the regulation of CVDs. For example, lncRNA LIPCAR is identified to be related to the severity of coronary artery disease (CAD) and used as a potential marker for heart failure (6,7). In addition, lncRNA UCA1 which is detectable in plasma, decreased in acute myocardial infarction (AMI) patients at 2 hours after the onset of symptoms (8). More importantly, our previous article (9,10) quantified that MIAT and H19 were increased in peripheral blood mononuclear cells (PBMCs) in AMI patients.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also named nuclear-enriched abundant transcript 2 (NEAT2), an 8,778 bp lncRNA, was found to be located at chromosome region 11q13 by Ji et al. (11) in patients with early-stage non-small-cell lung cancer (NSCLC). Among the prevalent lncRNAs, MALAT1 has attracted special attention, as it is obviously differentially expressed in cardiomyocytes and endothelial cells (ECs) under the risk conditions of hypoxia, oxidative stress and increased cytokines (12-14). Furthermore, MALAT1 was found to play a role in regulating cardiac development and in the pathogenesis of CVD. For example, Matkovich et al. (15)reported that MALAT1 played central roles in cardiac contractility, hypertrophy and failure, which correlated with miR-133. Although MALAT1 is highly conserved (16) in mammals, the single nucleotide polymorphisms (SNPs) were numerous within the region of lncRNA MALAT1 according to the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/). More studies have presented that SNPs in MALAT1 participate in lung cancer (17), hepatocellular carcinoma (18), colorectal cancer (19), and pulmonary arterial hypertension (20). Wang et al. (21) investigated the relationship between MALAT1 rs619586 and the potential risks of CAD and indicated that it could protect against the occurrence of CAD.
However, no data related to SNPs in MALAT1 and ACS risks have been reported. We carried out a retrospective case-control study to analyse the association between SNPs in MALAT1 and ACS risk in a Chinese population.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/cdt-20-906).
Methods
The protocol of this study was approved by the Human Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University (Urumqi, China) (approval ID: 20141201-03) and complied with the standards of the Declaration of Helsinki (22) (as revised in 2013). All participants provided written informed consent.
Study design and population
This single-centre, retrospective case-control study was designed to evaluate whether variants of the MALAT1 gene predispose patients to ACS. We consecutively recruited adult patients with ACS who attended the First Affiliated Hospital of Xinjiang Medical University from 2014 to 2018. Total ACS patients experienced chest pain within 12 h before undergoing percutaneous coronary intervention (PCI). The diagnostic criteria of case patients complied with the 2012 ACCF/AHA guidelines for ACS (23). The controls patients were consecutively recruited from the cardiovascular risk survey study in our hospital at the same time period, and they had no records of CAD based on the details of physical examination, ECG, blood tests, radiographic examination or coronary angiography. For all the subjects, the exclusion criteria were as follows: (I) thrombolysis therapy and other heart diseases (such as congenital heart disease, cardiomyopathy or severe valvular abnormalities); (II) active and chronic inflammation; and (III) renal or hepatic dysfunction, autoimmune diseases or malignancy. All the recruited participants were from Urumqi, excluding the surrounding and prefecture area. The design scheme of the study has been previously reported (24).
Blood collection and DNA extraction
For ACS patients, the time point of collection was strictly guaranteed to be within 12 hours after chest pain, before coronary angiography. Blood samples must be collected at the first time when the patient was admitted to the emergency department. Then, placed in EDTA-containing tubes immediately. To avoid affecting gene expression, samples must be separated and frozen within 2 hours. As previously described (25), DNA was extracted from peripheral blood leukocytes using a DNA extraction kit (Beijing Bioteke Corporation, Beijing, China). To make the subsequent experiments more convenient, DNA samples were diluted to 50 ng/µL and stored at –80 °C until use. We also randomly selected 92 ACS patients and 92 healthy controls to test the relative expression of lncRNA MALAT1. Total RNA was extracted by TRIzol reagent (Invitrogen), then detected the absorbance at 260 and 280 nm (A260/A280) by NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). If A260/A280 was between 1.8 and 2.1, 11 μL purified RNA was reverse transcription using the Qiagen Kit (Qiagen, China), then measured by quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR Green Master Mix (Applied Biosystems, USA) according to the manufacturer’s specifications at Xinjiang Key Laboratory of Cardiovascular Disease Research.
Biochemical analysis
All blood biochemical analyses were performed using a commercially automated biochemical analyser (Dimension AR/AVL Clinical Chemistry System, Newark, NJ, USA) in the Central Laboratory of the First Affiliated Hospital of Xinjiang Medical University. The tests included total triglyceride (TG), total cholesterol (TC), low-density lipoprotein-cholesterol (LDL-C) and high-density lipoprotein-cholesterol (HDL-C).
SNP selection
Using the HapMap human SNP database (www.hapmap.org) and the 1000G database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/), we received 12 SNPs in the 10 kb region of the MALAT1 gene based on the standard of minor allele frequency greater than 0.1 (with r2≥0.8 as a cut-off in linkage disequilibrium pattern analysis). Based on the tagged SNPs in the Chinese Han population and published references (19,21), we finally selected three SNPs (rs3200401, rs4102217, rs600231) in MALAT1.
Genotyping
Three sites of MALAT1 polymorphisms (rs3200401, rs4102217, rs600231) were genotyped using SNPscanTM typing assays with double ligation and multiplex fluorescence PCR (Cat#: G0104; Genesky Biotechnologies Inc., Shanghai, China). The technology was assisted via the Center for Genetic and Genomic Analysis. To ensure success rates of the genotyping in the study, repeat genotyping was performed in a random 5% of the sample, and the concordance rates of SNPs were more than 99%. The probe sequences of MALAT1 SNPs are shown in Table S1. In our study, the design scheme has been previously reported (26).
Definition of cardiovascular risk factors
Cardiovascular risk factors were defined according to current national guidelines. Hypertension (27) was defined as a history of hypertension or antihypertensive drug use, and/or systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg among repeated measurements. Diabetes (28) was diagnosed as fasting plasma glucose (FPG) ≥7.0 mmol/L and/or a history of antidiabetic drug use. Current smokers were considered those who reported tobacco use in the previous 6 months. Dyslipidaemia (29) was defined according to Chinese dyslipidaemia guidelines, and TG ≥1.71 mmol/L, TC ≥5.20 mmol/L, LDL-C ≥3.40 mmol/L, and HDL-C <1.04 mmol/L were defined as hypertriglyceridemia, hypercholesterolemia, hyper-LDL-C and low HDL-C, respectively.
Predictive nomogram for ACS
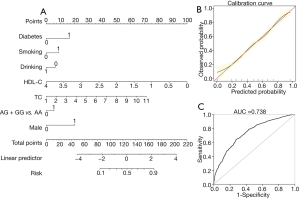
The predictive nomogram model for ACS was based on the results of multivariate logistic regression analysis and Akaike information criterion (AIC), including risk genotype and clinical variables. An individual patient’s value was located on each variable axis, and a line was drawn upward to determine the number of points received for each variable value. The sum of these numbers was located on the “total points” axis, and a line was drawn downward to the “risk” axis to determine the likelihood of ACS presence.
Statistical analysis
IBM SPSS Statistics 22.0 software (Chicago, IL, USA) was used for data analysis. Continuous variables are expressed as the mean ± standard deviation (SD) or median (quartile) and were compared using an independent-sample t-test or nonparametric rank test between ACS patients and controls. Numbers and percentages (%) were used to present the categorical variables, including the distribution of genotypes and models, and were compared using the chi-square test or Fisher’s exact test. Hardy-Weinberg equilibrium (HWE) was calculated with online SNP Stats (available online at http://bioinfo.iconcologia.net/SNPstats) to compare ACS patients and control subjects.
Logistic regression analyses were performed to assess the association between MALAT1 gene polymorphisms and the risk of ACS. The predictive nomogram model was built based on the stepwise regression screening of independent variables. Odds ratios (ORs) were calculated along with 95% confidence intervals (CIs). Statistical significance was set at P<0.05.
Results
Clinical characteristics of study participants
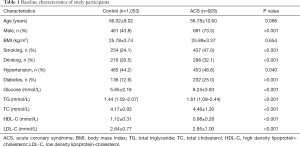
Table 1 shows the clinical characteristics of all study participants. In the study, 929 ACS patients (73.3% men and mean age 56.78±10.60 years) and 1,053 healthy controls (43.8% men and mean age 56.02±9.02 years) were recruited. There was no significant difference in age or body mass index (BMI) between the controls and ACS patients, which indicated that the study was an age-matched case-control study. In the ACS patients, the prevalence of diabetes and hypertension, the status of smoking and alcohol consumption, and the levels of glucose, TC, TG and LDL-C, were significantly higher compared to control subjects (P<0.05). However, the level of HDL-C was lower than that in the controls (P<0.05).
Full table
The genotype distribution of selected MALAT1 SNPs in ACS and controls
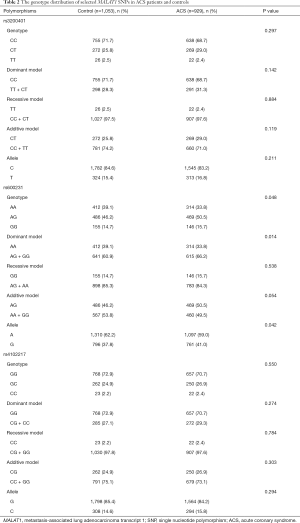
The distribution of alleles and genotypes of the selected SNPs of MALAT1 (rs3200401, rs600231, rs4102217) in all ACS patients and healthy controls are showed in Table 2. The distribution of genotype or allele rates in rs3200401 and rs4102217 showed no significant difference between control and ACS groups (P>0.05). Nevertheless, compared to control subjects, the frequency of the GG genotype was higher in ACS patients (15.7% vs. 14.7%, P=0.048). The dominant model of rs600231 (AG + GG vs. AA) was significantly different between ACS and control participants (66.2% vs. 60.9%, P=0.014). In ACS patients, the frequency of the G allele was higher than that in controls (41.0% vs. 37.8%, P=0.042).
Full table
The MALAT1 gene polymorphism rs600231 was positively associated with ACS
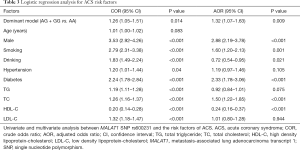
Univariate regression analysis presented that the dominant model of rs600231 was a risk factor for ACS (AG + GG vs. AA, OR =1.26, 95% CI: 1.05–1.51, P=0.014) (Table 3).Furthermore, multivariate logistic regression analysis was conducted to evaluate the relationship between susceptibility to ACS and rs600231 polymorphisms. After adjusting for confounding factors of ACS, such as sex, smoking, alcohol consumption, hypertension, diabetes and the plasma levels of TG, TC, LDL-C and HDL-C, the dominant model of rs600231 remained an independent risk factor for ACS (AG + GG vs. AA, OR =1.32, 95% CI: 1.07–1.63, P=0.009). However, there was no difference in the other two SNPs.
Full table
Predictive nomogram for ACS
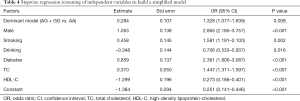
Based on their risk factors of the individuals, we add up all the scores to calculate their total score. The model was as follows: [–1.384+ (0.284× rs600231 AG + GG genotype) + (0.859× diabetes) + (0.458× smoking) + (0.370× TC) + (1.053× gender) – (0.348× alcohol consumption) – (1.299 × HDL-C)] (Table 4). As shown in Figure 1A, the score of each risk factor for ACS is listed in the nomogram. The calibration curve displayed in Figure 1B indicates good calibration of the nomogram. The area under the receiver operating characteristic (ROC) curve (AUC) for the nomogram model for the prediction of ACS was 0.738 (95% CI: 0.716–0.761) (Figure 1C). The proposed nomograms can be used for the prediction of ACS in this population.
Full table
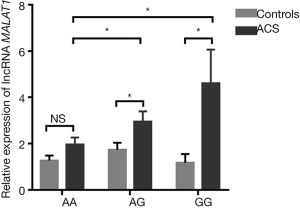
The expression of lncRNA MALAT1 was significantly associated with MALAT1 gene rs600231 polymorphisms
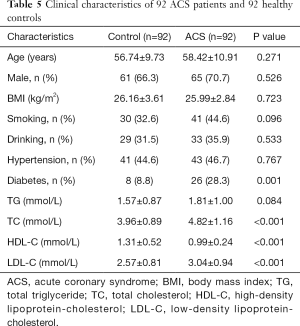
The relationship between lncRNA MALAT1 levels and MALAT1 gene rs600231 polymorphisms (AA, AG and GG genotypes) was further evaluated. In the AA genotype, no significant differences were found in lncRNA MALAT1 between controls and ACS patients (P>0.05). However, in AG and GG phenotypes, the relative expression of lncRNA MALAT1 was significantly higher in ACS patients than in controls carrying the same phenotypes (P<0.05). More importantly, the relative expression varied in the ACS patients carrying different genotypes: the highest was in those with the GG genotype, followed by those with the AG genotype, and the lowest was in those with the AA genotype (P<0.05) (Figure 2). The clinical characteristics of 92 healthy controls and 92 ACS patients are shown in Table 5.
Full table
Discussion
In the present case-control study, it was worth mentioning that we found a link between MALAT1 genetic variants (rs3200401, rs4102217, rs600231) and ACS in the Xinjiang population. Xinjiang, located in the northwest part of China, is a multi-ethnic co-populated region. It is well known that the lifestyles and habits of the Xinjiang population vary compared to those of populations in other regions of China. In the Chinese Han population in Xinjiang, our team’s previous studies have shown that the prevalence of hypertension, diabetes, dyslipidaemia and obesity was 33.4%, 9.26%, 53.3% and 18.8%, respectively, which was higher than that in other regions (30-34). After a multivariate adjustment analysis for potential confounding factors, there was a significant difference between the polymorphism (rs600231) of MALAT1 gene and ACS. To our knowledge, this was the first study of the relationship between the MALAT1 gene and ACS patients in Xinjiang, China.
Atherosclerosis, inflammation and vascular dysfunctions are the main risk factors for coronary heart disease, including in the pathogenesis of ACS (35-37). Previous studies (38,39) have reported that MALAT1 not only participates in the development of atherosclerosis but also indicates a relationship with endothelial dysfunction. Yan et al. (40,41) reported that MALAT1, through improving CD36 expression induced by ox-LDL in macrophages or enhancing the stability of SREBP-1C protein, could increase hepatic lipid accumulation. Several studies (42,43) documented that the relative level of MALAT1 was higher in high-fat food-fed ApoE–/– mice treated with ox-LDL, which could enhance the effect of EndMT and activate Wnt/β-catenin signalling. Inflammatory cytokines may also induce ECs injury by oxidative damage, and MALAT1 not only enhances oxidative stress in ECs but can also be upregulated by IL-6 via the mechanism of an extracellular regulated protein kinases (ERK) signal (44). In addition, MALAT1 was increased in a hypoxia-reoxygenation (H/R) cell or ischaemia/reperfusion (I/R) mouse model and could inhibit the expression of PTEN by sponging miR-320 in various physiological processes, including cell migration, differentiation and apoptosis (45). Considering the underlying biological function, we analysed the effects about SNPs in MALAT1 among ACS patients.
In our study, the result showed that rs600231 AG and GG genotypes carriers were more susceptible to ACS than AA genotypes carriers. The dominant model (AG + GG vs. AA) and G allele of rs600231 presented distinct differences among ACS patients and controls (P<0.05). After adjusted the covariates, including age, sex, hypertension, diabetes, smoking, alcohol consumption, TC, TGs, HDL-C and HDL-C, the significant difference in the polymorphism (rs600231) of the MALAT1 gene and ACS remained. These findings suggested that rs600231 in MALAT1 is a risk factor for ACS (AG + GG vs. AA, OR =1.32, 95% CI: 1.07–1.63, P<0.05). Furthermore, using above clinical variables and rs600231 of MALAT1, we firstly developed a diagnostic nomogram model which was of high discrimination and calibration to predict the risk of ACS. The calibration curve displayed in nomogram model indicates good calibration, which the area under ROC curve for the evaluating of ACS was 0.738 (95% CI: 0.716–0.761). MALAT1 rs600231 was selected because of its regulation of gene expression, which could remotely control the promoter region (46-48). However, rs600231 was explored in the context of its association with cancer, and we found no related results for CAD. Previous studies have also explored whether rs3200401 and rs4102217 of MALAT1 are involved in the prediction of CAD. Similar findings (49,50) that the two SNPs were not associated with the onset of CAD were reported, but these studies recognized the linking with the CAD risk factors, such as diabetes, the status of smoking and lipids (TC and TG).
In addition, the relative expression of the lncRNA MALAT1 was measured in PBMCs from parts of controls and ACS patients. We revealed that enrolled participants carrying GG phenotype demonstrated the highest lncRNA MALAT1 expression. The relative expression of lncRNA MALAT1 in ACS who carried AG or GG phenotype was obvious higher than that in controls who carried the same phenotypes, but no difference was found in the AA phenotype. The first study (51) on circulating lncRNAs in 414 AMI patients and 86 healthy controls tested the relative expression of lncRNA MALAT1. As a result, it was obvious that the lncRNA MALAT1 was more highly expressed in AMI patients than in controls. Indeed, in 132 AMI patients and 104 controls, our previous research (9) found that the relative expression of lncRNA MALAT1 was higher in AMI patients than in controls.
However, there are some limitations to our study. First, our present study evaluated only the relationship between three SNPs of the MALAT1 gene (rs3200401, rs4102217 and rs600231) and ACS risks among the Chinese population in the Xinjiang region, and the other SNPs of the MALAT1 gene were not evaluated. Second, the enrolled ACS subjects in our present study were from one hospital because this was not a multicentre large-scale study, and thus, the sample size limitation and selection bias could not be avoided. Finally, we measured only the lncRNA MALAT1 in a small portion of unmatched participants, so the analysis might not have had enough power to determine the real effect.
In conclusion, our study provides the first evidence between the risk factor of rs600231 in MALAT1 gene and the susceptibility of ACS in the Chinese population (Xinjiang). MALAT1 gene polymorphisms may present predictive value for the risk of ACS patients, while the combination of MALAT1 gene polymorphisms (AG and GG genotypes) and the expression of lncRNA MALAT1 may improve the ability to predict the severity of ACS. Further studies also need to be performed to elucidate the potential molecular mechanism of rs600231 in the MALAT1 gene in ACS.
Acknowledgments
Funding: This research was funded by the Major Disease Medical Key Laboratory Open Subject of Xinjiang in China (2018D04029), the National Natural Science Foundation of China (81660058, 81770363), Natural Science Foundation of the Xinjiang Uygur Autonomous Region in China (2017D01C362) and the Scientific research project in Xinjiang Medical University (No. 33-0104006020801).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/cdt-20-906
Data Sharing Statement: Available at http://dx.doi.org/10.21037/cdt-20-906
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-906). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Human Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University (Urumqi, China) (approval ID: 20141201-03). All participants provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation 2017;135:e146-603. Erratum in: Circulation 2017;135:e646 Erratum in: Circulation 2017;136:e196. [Crossref] [PubMed]
- Shibata T, Kawakami S, Noguchi T, et al. Prevalence, clinical features, and prognosis of acute myocardial infarction attributable to coronary artery embolism. Circulation 2015;132:241-50. [Crossref] [PubMed]
- Zhang H, Masoudi FA, Li J, et al. National assessment of early β-blocker therapy in patients with acute myocardial infarction in China, 2001-2011: The China Patient-centered Evaluative Assessment of Cardiac Events (PEACE)-Retrospective AMI Study. Am Heart J 2015;170:506-15.e1. [Crossref] [PubMed]
- Hartman MHT, Eppinga RN, Vlaar PJJ, et al. The contemporary value of peak creatine kinase-MB after ST-segment elevation myocardial infarction above other clinical and angiographic characteristics in predicting infarct size, left ventricular ejection fraction, and mortality. Clin Cardiol 2017;40:322-8. [Crossref] [PubMed]
- Jaffe AS, Babuin L, Apple FS. Biomarkers in acute cardiac disease: the present and the future. J Am Coll Cardiol 2006;48:1-11. [Crossref] [PubMed]
- Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014;114:1569-75. [Crossref] [PubMed]
- Li M, Wang YF, Yang XC, et al. Circulating long noncoding RNA LIPCAR acts as a novel biomarker in patients with ST-segment elevation myocardial infarction. Med Sci Monit 2018;24:5064-70. [Crossref] [PubMed]
- Yan Y, Zhang B, Liu N, et al. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int 2016;2016:8079372 [Crossref] [PubMed]
- Wang XM, Li XM, Song N, et al. Long non-coding RNAs H19, MALAT1 and MIAT as potential novel biomarkers for diagnosis of acute myocardial infarction. Biomed Pharmacother 2019;118:109208 [Crossref] [PubMed]
- Ishii N, Ozaki K, Sato H, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087-99. [Crossref] [PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. [Crossref] [PubMed]
- Zeng R, Zhang R, Song X, et al. The long non-coding RNA MALAT1 activates Nrf2 signaling to protect human umbilical vein endothelial cells from hydrogen peroxide. Biochem Biophys Res Commun 2018;495:2532-8. [Crossref] [PubMed]
- Zhao G, Su Z, Song D, et al. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS Lett 2016;590:2884-95. [Crossref] [PubMed]
- Wang LQ, Zhou HJ. LncRNA MALAT1 promotes high glucose-induced inflammatory response of microglial cells via provoking MyD88/IRAK1/TRAF6 signaling. Sci Rep 2018;8:8346. [Crossref] [PubMed]
- Matkovich SJ, Wang W, Tu Y, et al. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res 2010;106:166-75. [Crossref] [PubMed]
- Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [Crossref] [PubMed]
- Gong WJ, Peng JB, Yin JY, et al. Association between well-characterized lung cancer lncRNA polymorphisms and platinum-based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacol Sin 2017;38:581-90. [Crossref] [PubMed]
- Liu Y, Pan S, Liu L, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One 2012;7:e35145 [Crossref] [PubMed]
- Li Y, Bao C, Gu S, et al. Associations between novel genetic variants in the promoter region of MALAT1 and risk of colorectal cancer. Oncotarget 2017;8:92604-14. [Crossref] [PubMed]
- Zhuo Y, Zeng Q, Zhang P, et al. Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin Chem Lab Med 2017;55:38-46. [Crossref] [PubMed]
- Wang G, Li Y, Peng Y, et al. Association of polymorphisms in MALAT1 with risk of coronary atherosclerotic heart disease in a Chinese population. Lipids Health Dis 2018;17:75. [Crossref] [PubMed]
- World Medical Association. Declaration of Helsinki. J Am Med Assoc 2013;227:925-6.
- Jneid H, Anderson JL, Wright RS, et al. 2012 ACCF/AHA focused update of the guideline for the management of patients with unstable angina/non-ST-elevation myocardial infarction (updating the 2007 guideline and replacing the 2011 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 2012;60:645-81. [Crossref] [PubMed]
- Du GL, Luo JY, Wang D, et al. MIF gene rs755622 polymorphism positively associated with acute coronary syndrome in Chinese Han population: case-control study. Sci Rep 2020;10:140. [Crossref] [PubMed]
- An Y, Wang YT, Ma YT, et al. IL-10 genetic polymorphisms were associated with valvular calcification in Han, Uygur and Kazak populations in Xinjiang, China. PLoS One 2015;10:e0128965 [Crossref] [PubMed]
- Chen X, Li S, Yang Y, et al. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J Thromb Haemost 2012;10:1508-14. [Crossref] [PubMed]
- Jones DW, Hall JE. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure and evidence from new hypertension trials. Hypertension 2004;43:1-3. [Crossref] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2011;34:S62-9. [Crossref] [PubMed]
- . Joint Committee for Developing Chinese guidelines on Prevention and Treatment of Dyslipidemia in Adults. Chinese guidelines on prevention and treatment of dyslipidemia in adults. Zhonghua Xin Xue Guan Bing Za Zhi 2007;35:390-419.
- Liu F, Ma YT, Yang YN, et al. Current status of primary hypertension in Xinjiang:an epidemiological study of Han, Uygur and Hazakh populations. Zhonghua Yi Xue Za Zhi 2010;90:3259-63. [PubMed]
- Ma LY, Chen WW, Gao RL, et al. China cardiovascular diseases report 2018: an updated summary. J Geriatr Cardiol 2020;17:1-8. [PubMed]
- Yang YN, Xie X, Ma YT, et al. Type 2 diabetes in Xinjiang Uygur autonomous region, China. PLoS One 2012;7:e35270 [Crossref] [PubMed]
- Tao J, Ma YT, Xiang Y, et al. Prevalence of major cardiovascular risk factors and adverse risk profiles among three ethnic groups in the Xinjiang Uygur Autonomous Region, China. Lipids Health Dis 2013;12:185. [Crossref] [PubMed]
- Song N, Liu F, Han M, et al. Prevalence of overweight and obesity and associated risk factors among adult residents of northwest China: a cross-sectional study. BMJ Open 2019;9:e028131 [Crossref] [PubMed]
- Leong DP, Joseph PG, McKee M, et al. Reducing the global burden of cardiovascular disease, part 2: prevention and treatment of cardiovascular disease. Circ Res 2017;121:695-710. [Crossref] [PubMed]
- Puthanveetil P, Chen S, Feng B, et al. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med 2015;19:1418-25. [Crossref] [PubMed]
- Yan Y, Song D, Song X, et al. The role of lncRNA MALAT1 in cardiovascular disease. IUBMB Life 2020;72:334-42. [Crossref] [PubMed]
- Cremer S, Michalik KM, Fischer A, et al. Hematopoietic deficiency of the long noncoding RNA MALAT1 promotes atherosclerosis and plaque inflammation. Circulation 2019;139:1320-34. [Crossref] [PubMed]
- Gast M, Rauch BH, Nakagawa S, et al. Immune system-mediated atherosclerosis caused by deficiency of long non-coding RNA MALAT1 in ApoE-/-mice. Cardiovasc Res 2019;115:302-14. [Crossref] [PubMed]
- Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep 2016;6:22640. [Crossref] [PubMed]
- Huangfu N, Xu Z, Zheng W, et al. LncRNA MALAT1 regulates oxLDL-induced CD36 expression via activating beta-catenin. Biochem Biophys Res Commun 2018;495:2111-7. [Crossref] [PubMed]
- Li H, Zhao Q, Chang L, et al. LncRNA MALAT1 modulates ox-LDL induced EndMT through the Wnt/beta-catenin signaling pathway. Lipids Health Dis 2019;18:62. [Crossref] [PubMed]
- Vanhoutte PM, Shimokawa H, Tang EH, et al. Endothelial dysfunction and vascular disease. Acta Physiol (Oxf) 2009;196:193-222. [Crossref] [PubMed]
- Wang Y, Nie W, Yao K, et al. Interleukin 6 induces expression of NADPH oxidase 2 in human aortic endothelial cells via long noncoding RNA MALAT1. Pharmazie 2016;71:592-7. [PubMed]
- Hu H, Wu J, Li D, et al. Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed Pharmacother 2018;106:738-46. [Crossref] [PubMed]
- Bacci L, Barbati SA, Colussi C, et al. Sildenafil normalizes MALAT1 level in diabetic cardiomyopathy. Endocrine 2018;62:259-62. [Crossref] [PubMed]
- Abdulle LE, Hao JL, Pant OP, et al. MALAT1 as a Diagnostic and therapeutic target in diabetes-related complications: a promising long-noncoding RNA. Int J Med Sci 2019;16:548-55. [Crossref] [PubMed]
- Zhang D, Cheng L, Badner JA, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet 2010;86:411-9. [Crossref] [PubMed]
- Li Q, Zhu W, Zhang B, et al. The MALAT1 gene polymorphism and its relationship with the onset of congenital heart disease in Chinese. Biosci Rep 2018;38:BSR20171381 [Crossref] [PubMed]
- Li Y, Zhang D, Zhang Y, et al. Association of lncRNA polymorphisms with triglyceride and total cholesterol levels among myocardial infarction patients in Chinese population. Gene 2020;724:143684 [Crossref] [PubMed]
- Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 2014;115:668-77. [Crossref] [PubMed]


