
Association between CDK8 gene polymorphisms and dilated cardiomyopathy in a Chinese Han population
Introduction
Dilated cardiomyopathy (DCM) is a primary myocardial disease defined by dilation of the left or both ventricles and contractile dysfunction, with decreased ejection fraction (1). DCM is also the most common type of cardiomyopathy, with a prevalence rate of 1/2,500 (2). The manifestations of DCM include progressive heart failure, ventricular arrhythmia, and sudden cardiac death (3). DCM is one of the leading causes of death in Western developed countries (4). The etiology of DCM can be divided into non-genetic and genetic (5). In previous research, thousands of mutations in more than 50 genes have been reported to be related to the pathogenesis of familial DCM, such as ACTC1, TTN, LAMA4, and SCN5A, among others (3). Most of these genes define heart structures and cardiac functions, but they can only explain 40% of the pathogenesis of DCM, since DCM is genetically heterogeneous (6). At present, the mechanism of gene mutation in the pathogenesis of DCM remains partially unclear. Accumulating studies have demonstrated that some transcription factors (TFs) are significantly associated with the dysfunction of cardiac muscles, particularly under pathological stress. Additionally, some studies have raised the possibility of cyclin-dependent kinase 8 (CDK8) variants as potential risk factors of some cardiomyopathies (7,8).
The CDK8 gene encodes a major isoform of 464 amino acids and is an evolutionarily conserved serine/threonine kinase component of the Module (8). The Module is a 4-subunit complex that binds to the Mediator and regulates its activity. The Mediator complex is a global regulator of gene expression through the phosphorylation of RNA polymerase II and is considered as a general TF. Thus, CDK8 has a vital role in regulating transcription processes. Meanwhile, CDK8 activity is important in the expression of numerous components of the glycolytic cascade (9) and as a key regulator of several additional signaling pathways, such as the WNT, TGFβ/BMP, STAT1, SHH, and NOTCH pathways (10,11). Some researchers recently reported that CDK8-related disorders present characteristic facial features, congenital heart disorders, and intellectual disability in humans (7,8). Overexpression of CDK8 causes progressive heart failure, DCM, and premature lethality (12). The basic mechanisms of CDK8-related disorders have yet to be delineated.
Moreover, single nucleotide polymorphisms (SNPs) of CDK8 have not yet been revealed as predictors or risk factors of DCM prognosis and susceptibility. This study firstly hypothesized that SNPs of the CDK8 gene might affect the susceptibility and prognosis of DCM. A pilot study was designed to investigate the role of CDK8 polymorphisms among DCM patients in a Chinese Han population. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/cdt-21-323).
Methods
Study subjects
The case group contained a total of 341 unrelated patients diagnosed with DCM (mean age: 49.37±14.432; gender: male, 226; female, 115) recruited from West China Hospital of Sichuan University from July 2005 to August 2019. These patients were diagnosed with DCM according to the criteria established by the World Health Organization/International Society and Federation of Cardiology Task Force on the Classification of Cardiomyopathies in 1995 (before 2006) (13), the Scientific Statement on the Classification of Cardiomyopathies proposed by the American Heart Association in 2006 (2006–2008) and the Classification from the European Society of Cardiology in 2008 (after 2008) (14). Cardiac function was graded by the New York Heart Association (NYHA) classification, and 72 patients were NYHA II, 207 patients were NYHA III, and 62 patients were NYHA IV. All patients were treated under the guidance of clinical guidelines in this study. Echocardiography was used to assess heart function. A total of 381 healthy unrelated individuals (mean age: 48.98±16.772; gender: male, 234; female, 147) from routine health surveys were recruited as the control group, with NYHA functional classes ranging from I to II. Age and gender in the control group were matched with the case group. Individuals with a history of coronary heart disease, cardiac valve disease, hypertension, tachyarrhythmia, acute viral myocarditis, familial DCM history, heavy alcohol intake, diabetes, obesity, insulin resistance, systemic diseases of putative autoimmune origin, skeletal myopathies, Parkinson’s disease, or Alzheimer’s diseases were excluded intentionally. Clinical data were collected from the hospital record system. We did not detect whether these patients had the most frequent monogenic causes of DCM, and all subjects were from the Chinese Han population of Sichuan Province in southwestern China. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This research was approved by the Hospital Ethics Committee (No. 81670346) and written informed consents from all subjects were obtained.
SNP selection and genotyping
EDTA-anticoagulated peripheral blood samples (5 mL) were drawn from each participant. A DNA isolation kit (BioTeke, Peking, China) was used according to the manufacturer’s protocol to extract genomic DNA from 200 µL whole blood samples (15). These DNA products were stably stored in a −80 °C freezer until assayed.
Tag SNPs were selected based on the data of the Chinese Han in Beijing China population sample from the HapMap Project (Data Release 24/Phase II, NCBI build 36 assembly, dpSNPb126). Two tag SNPs of CDK8, rs17083838 (A/G) and rs7992670 (A/G) were selected by the National Institute of Environmental Health Sciences (NIEHS) online software. PCR primers were designed using the Primer 3 software (https://primer3.ut.ee/) (16).
CDK8 gene polymorphisms were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). PCR reactions were performed in a 10 µL reaction system containing 50 ng genomic DNA, 0.15 µL forward and reverse primers, 5 µL 2× Power Taq PCR Master Mix (Biosharp, Peking, China), and 10 µL ddH2O. The PCR conditions were an initial denaturation step at 94 °C for 2 min, 34 cycles at 94 °C for 30 s, annealing at 58 °C for 30 s (rs17083838 and rs7992670), extension at 72 °C for 30 s, and final elongation at 72 °C for 10 min. PCR products were digested stably by the specific restriction enzymes MspI for 45 min (rs17083838) and Mboll for 30 min (rs7992670) at 37 °C according to the manufacturer’s instructions (New England Biolabs, Peking, China) (Table 1). Ultimately, the digested products were electrophoresed and visualized by 6% polyacrylamide gels with silver staining. A double-blind method was applied during the genotyping of the enrolled subjects. Although DNA sequencing is the most accurate choice for SNPs genotyping, PCR and agarose gel electrophoresis were still used in our study as they are convenient and fast. To verify the accuracy of the genotyping results, ~10% PCR-amplified DNA samples randomly underwent repeated visualization, and all results were 100% consistent.
Table 1
| SNP ID | Primer sequence | Annealing temperature (°C) | Restriction enzyme | Allele (bp) |
|---|---|---|---|---|
| rs17083838 | F: 5'-TCTGTTGCACTTGCCATATCA-3' | 58 | MspI | A [194] |
| R: 5'-TCCAAAGACTTTCAAAGACACTCA-3' | G [194=68+126] | |||
| rs7992670 | F: 5'-TTTCATGCAGATACCACGTCA-3' | 58 | MboII | A [151] |
| R: 5'-TGGATTCACGGGTTCGTTAT-3' | G [151=113+38] |
bp, base pair; SNP, single nucleotide polymorphism.
Clinical follow-up
A total of 153 patients with contact information were scheduled for follow-up every 3 months. The basic clinical characteristics (gender, age, blood pressures and heart rate) of patients and echocardiographic indicators [left ventricular end-diastolic diameter (LVEDD) and left ventricular ejection fraction (LVEF)] were obtained from medical records, and echocardiography was performed at the time of diagnosis using an S5-1 broadband phased-array transducer (1–5 MHz) (Philips Medical Systems, Bothell, WA, USA). The end points were cardiac death or loss to follow-up. During follow-up, all patients received continuous medication treatment, and no patients underwent heart transplantation. Follow-up was performed in a double-blind manner, and the genotype of patients remained unknown.
Statistical analysis
Data analysis was performed using SPSS version 26.0 software (SPSS Inc., Chicago, IL, USA). Power and Sample Size Calculation software (version 3.0.43) was used to calculate sample power (17). Differences between two independent samples of continuous data were analyzed using the Mann-Whitney U test, while categorical variables were analyzed with Pearson’s chi-square test. Shapiro-Wilk’s test was used to determine normality. Genotypic frequencies of SNPs were obtained by counting directly. To compare the allelic and genotypic distribution between the DCM and healthy control groups, data were evaluated on the SNPStats website in the codominant, dominant, recessive, and overdominant models (18). The chi-square test was used to evaluate the Hardy-Weinberg equilibrium (HWE). Linkage disequilibrium tests and haplotype analysis were carried out using SHEsis software (19,20). The odds ratio (OR) and its 95% confidence interval (CI) were calculated to assess the effects of any differences in the distribution of genotypes and alleles. For survival analyses, Kaplan-Meier curve analysis and Cox regression were performed. A P value <0.05 was considered statistically significant in all the statistical analyses. Bonferroni corrections were carried out for multiple testing to determine statistical significance by the number of analyzed SNPs (for 2 SNPs, a Bonferroni correction coefficient of 2 was applied to the P values).
Results
Baseline characteristics of the enrolled subjects
The baseline clinical and echocardiographic characteristics of all participants are summarized in Table 2. The DCM group had a lower systolic blood pressure, diastolic blood pressure and LVEF than the control group. The HR and LVEDD of DCM patients were higher than those of control subjects (both P<0.001). Age and gender were matched between the two groups (P=0.737 and P=0.175 respectively). All samples had >80% power, which revealed that the sample size was adequate in the present study.
Table 2
| Variables | Cases (n=341) | Controls (n=381) |
|---|---|---|
| Age (years) | 49.37±14.432 | 48.98±16.772 |
| Gender (male/female) | 226/115 | 234/147 |
| NYHA | II: 72; III: 207; IV: 62 | I: 306; II: 75 |
| SBP (mmHg) | 108.62±17.681* | 114.17±12.672 |
| DBP (mmHg) | 61.62±9.453* | 74.44±8.410 |
| HR (beats/min) | 91.71±12.197* | 80.62±11.425 |
| LVEDD (mm) | 67.79±8.987* | 47.11±5.198 |
| LVEF (%) | 32.07±11.023* | 62.98±7.188 |
Data are presented as the mean ± SD or number. *, patients
Associations between CDK8 SNPs and the susceptibility to DCM
Two SNPs of the CDK8 gene, rs17083838 (A/G) and rs7992670 (A/G), were previously identified by the International HapMap project (https://www.genome.gov/10001688/international-hapmap-project). The allelic and genotypic distribution of these 2 SNPs were consistent with the expected HWE (P=0.81 for rs17083838; P=0.55 for rs7992670), suggesting that all the samples met the assumption of the HWE (P>0.05). To identify the association between susceptibility to DCM and the two SNPs, a chi-square test was performed by SNPStats to analyze the genotype and allele frequencies in the DCM and control groups. Significant differences concerning both SNPs are summarized in Table 3.
Table 3
| Model | Genotype | Cases, n (%) | Controls, n (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| rs17083838 | |||||
| Codominant | GG | 210 (61.6) | 259 (61.9) | 1.00 | – |
| AG | 110 (32.4) | 110 (29.3) | 1.23 (0.89–1.70) | – | |
| AA | 21 (6.2) | 6 (1.6) | 3.96 (1.05–1.94)* | 0.0021* | |
| Dominant | GG | 210 (61.6) | 259 (69.1) | 1.00 | – |
| AG-AA | 131 (38.4) | 116 (30.9) | 1.39 (1.02–1.90)* | 0.035* | |
| Recessive | GG-AG | 320 (93.8) | 369 (98.4) | 1.00 | – |
| AA | 21 (6.2) | 6 (1.6) | 4.04 (1.61–10.12)* | 0.0011* | |
| Overdominant | GG-AA | 231 (67.7) | 265 (70.7) | 1.00 | – |
| AG | 110 (32.3) | 110 (29.3) | 1.15 (0.83–1.58) | 0.4 | |
| Allele | G | 530 (78.0) | 628 (84.0) | 1.00 | – |
| A | 152 (22.0) | 122 (16.0) | 0.677 (0.520–0.883) | 0.004* | |
| rs7992670 | |||||
| Codominant | GG | 85 (24.9) | 114 (30.9) | 1.00 | – |
| AG | 172 (50.4) | 189 (51.2) | 1.22 (0.86–1.73) | – | |
| AA | 84 (24.6) | 66 (17.9) | 1.71 (1.11–2.62)* | 0.047* | |
| Dominant | GG | 85 (24.9) | 114 (30.9) | 1.00 | – |
| AG-AA | 256 (75.1) | 255 (69.1) | 1.35 (0.97–1.87) | 0.076 | |
| Recessive | GG-AG | 257 (75.4) | 303 (82.1) | 1.00 | – |
| AA | 84 (24.6) | 66 (17.9) | 1.50 (1.04–2.16)* | 0.028* | |
| Overdominant | GG-AA | 169 (49.6) | 180 (48.8) | 1.00 | – |
| AG | 172 (50.4) | 189 (51.2) | 0.97 (0.72–1.30) | 0.84 | |
| Allele | G | 342 (50.0) | 417 (57.0) | 1.00 | – |
| A | 340 (50.0) | 321 (43.0) | 0.774 (0.628–0.954) | 0.016* | |
P<0.05 was considered to be statistically significant; *, values indicate statistical significance. SNP, single nucleotide polymorphism; DCM, dilated cardiomyopathy; CI, confidence interval; OR, odds ratio.
The results revealed that the frequency of the A allele in SNP rs17083838 among DCM patients was notably elevated compared with the control group (22.0% vs. 16.0%; P=0.004; OR: 0.677; 95% CI: 0.520–0.883). In the codominant model, the frequencies of the GG, AG, and AA genotypes of rs17083838 were, respectively, 61.6%, 32.4%, and 6.2% in DCM cases, and 61.9%, 29.3%, and 1.6% in controls. The differences among genotype frequencies were statistically significant (P=0.0021; OR: 3.96; 95% CI: 1.05–1.94). Associations between s17083838 and DCM susceptibility were also found in the dominant model, where A carriers (AA/AG genotypes) had a higher DCM risk than the GG genotype (P=0.035; OR: 1.39; 95% CI: 1.027–1.90), and in the recessive model, where the AA genotype had a higher DCM risk than the GG/AG genotypes (P=0.0011; OR: 4.04; 95% CI: 1.61–10.12). These results remained significant even after the Bonferroni correction (P<0.025).
For the allele frequencies of SNP rs7992670, the DCM group had a significantly greater allele frequency of A than the healthy control group (50% vs. 43%, P=0.016, OR: 0.774, 95% CI: 0.628–0.954). AA genotypes had higher DCM risk than AG/GG genotypes in the codominant model (P=0.047; OR: 1.71; 95% CI: 1.11–2.62). In the recessive model, compared with the GG/AG genotypes, an increased risk of DCM was associated with the AA genotype (P=0.028; OR: 1.50; 95% CI: 1.04–2.16). The association was still significant after the Bonferroni correction (P<0.025). However, no significant differences were observed in the dominant and overdominant models.
There were no obvious differences between DCM patients and healthy controls in the overdominant model for both rs17083838 (P=0.4, OR: 1.15, 95% CI: 0.83–1.58) and rs7992670 (P=0.84 OR: 0.97, 95% CI: 0.72–1.30). Hence, stratified analyses were conducted for this model among 341 DCM patients. Table 4 summarizes the results after adjusting for age, LVEF, and LVEDD. The frequency of the GG/AA genotype was higher in individuals with a larger left ventricle (LV ≥67 mm) than that in patients with LVEDD less than 67 mm for rs7992670 (57.4% vs. 45.1%, OR: 0.608, 95% CI: 0.396–0.935, P=0.023). No correlation was found between LVEDD and rs17083838, and there were no significant differences between the two SNPs (rs17083838 and rs7992670) and the other indicators (age, gender, and LVEF).
Table 4
| Characteristics | rs17083838 | rs7992670 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | OR (95% CI) | P | Genotype | OR (95% CI) | P | ||||
| GG-AA | AG | GG-AA | AG | ||||||
| Age (year), n (%) | |||||||||
| <50 | 111 (68.1) | 52 (31.9) | – | – | 90 (55.2) | 73 (44.8) | – | – | |
| ≥50 | 120 (67.4) | 58 (32.6) | 1.032 (0.655–1.626) | 0.893 | 87 (48.9) | 91 (51.1) | 1.290 (0.842–1.975) | 0.242 | |
| Gender, n (%) | |||||||||
| Male | 150 (66.4) | 76 (33.6) | – | – | 116 (51.3) | 110 (48.7) | – | – | |
| Female | 81 (70.4) | 34 (29.6) | 0.828 (0.509–1.347) | 0.448 | 61 (53.0) | 54 (47.0) | 0.934 (0.595–1.464) | 0.764 | |
| LVEDD (mm), n (%) | |||||||||
| <67 | 97 (63.4) | 56 (36.6) | – | – | 69 (45.1) | 84 (54.9) | – | – | |
| ≥67 | 134 (71.3) | 54 (28.7) | 0.698 (0.442–1.101) | 0.122 | 108 (57.4) | 80 (42.6) | 0.608 (0.396–0.935)* | 0.023* | |
| LVEF, n (%) | |||||||||
| <30 | 110 (70.5) | 46 (29.4) | – | – | 82 (52.6) | 74 (47.4) | – | – | |
| ≥30 | 121 (65.4) | 64 (34.6) | 1.265 (0.800–2.001) | 0.315 | 95 (51.4) | 90 (48.6) | 1.050 (0.685–1.608) | 0.823 | |
Adjusted for age, gender, LVEDD, and LVEF. *, values indicate statistical significance. SNP, single nucleotide polymorphism; DCM, dilated cardiomyopathy; OR, odds ratio.
Linkage disequilibrium and haplotype analysis
Linkage disequilibrium was observed within the two variants using SHEsis software (http://analysis.bio-x.cn/myAnalysis.php). The results showed that SNP rs17083838 and rs7992670 were not in linkage disequilibrium (D’=0.148, r2=0.004), which meant both of the SNPs were representative. We found a significant association in the distribution of the three haplotype frequencies (AA, AG, and GG) between DCM cases and healthy controls (P<0.05) (Table 5) in the further analysis of four haplotype combinations. The haplotype AA was significantly associated with increased DCM susceptibility, while GG was a protective haplotype.
Table 5
| CDK8 gene (rs17083838/rs7992670) haplotypes | Frequency | OR (95% CI) | P value | |
|---|---|---|---|---|
| Cases | Controls | |||
| A A | 61.59 (0.090) | 43.03 (0.059) | 1.576 (1.052–2.361) | 0.026518* |
| A G | 90.41 (0.133) | 70.97 (0.098) | 1.411 (1.014–1.962) | 0.040427* |
| G A | 278.41 (0.408) | 271.97 (0.375) | 1.152 (0.930–1.427) | 0.196346 |
| G G | 251.59 (0.369) | 340.03 (0.468) | 0.663 (0.536–0.821) | 0.000159* |
Haplotypes with frequency less than 3.0% were not analyzed; *, significant P value after multiple testing adjustments (P<0.05). DCM, dilated cardiomyopathy; OR, odd ratio; CI, confidence interval.
Survival analysis
Survival analysis was performed to reveal the correlation between DCM prognosis and the two SNPs (rs17083838 and rs7992670) of the CDK8 gene. In this study, we continuously tracked a total of 153 DCM patients with contact information (males/females, 106/46; mean age, 52.35±12.269 years) for a mean period of 55.36±23.170 months. All included DCM patients received continuous medication treatment during the follow-up period, and none of them underwent heart transplantation. Eighteen (11.8%) DCM patients were withdrawn due to loss to follow-up, 89 (58.2%) DCM patients died, and 46 (30.1%) were alive at the end of our follow-up.
Kaplan-Meier curves indicated that the AA genotype of CDK8 rs17083838 was associated with a worse prognosis for DCM patients in the dominant model (Log- rank: P=0.020, Figure 1). For rs7992670, the AA genotype was associated with a worse prognosis of DCM patients in the recessive model (log-rank: P<0.001, Figure 2). In the univariate Cox regression analysis, a positive correlation was found between the genotypic frequencies of rs17083838 and the overall survival (OS) of DCM patients in the dominant model (HR: 1.623, 95% CI: 1.069–2.464, P=0.023), with the A carriers (AA/AG) associated with a worse prognosis. Meanwhile, the AA genotype of rs7992670 in the recessive model had a worse prognosis among DCM patients (P=0.001; HR: 2.054; 95% CI: 1.347–3.133). These results were identical to those of the Kaplan-Meier analysis. The results remained statistically significant in the multivariate Cox analysis after adjusting for age, gender, LVEDD and LVEF (Table 6). Additionally, the Bonferroni correction was applied for multiplicity comparison. All the above data remained significant after the Bonferroni correction (P<0.025).
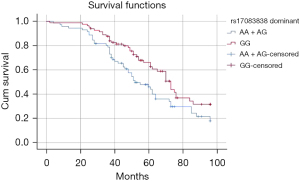
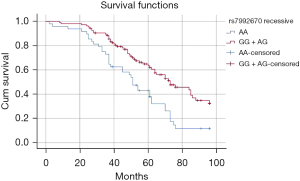
Table 6
| Characteristic model | Genotype | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate survival analysis | Multivariate survival analysis* | |||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |||
| rs17083838 | ||||||||
| Dominant | GG | 1.00 | – | – | 1.00 | – | – | |
| AA/AG | 1.623 | 1.069–2.464# | 0.023# | 1.539 | 1.013–2.340# | 0.043# | ||
| rs7992670 | ||||||||
| Recessive | GG/AG | 1.00 | – | – | 1.00 | – | – | |
| AA | 2.054 | 1.347–3.133# | 0.001# | 2.146 | 1.405–3.280# | <0.001# | ||
#, values indicate statistical significance; *, adjusted for gender, age, LVEDD, and LVEF. SNP, single nucleotide polymorphism; DCM, dilated cardiomyopathy; HR, hazard ratio; CI, confidence interval.
Discussion
The pathogenic mechanism of DCM remains unclear and is a research hotspot in cardiology. Numerous genes have been reported to be related to the susceptibility and prognosis of DCM. Determining the exact gene responsible for DCM is necessary on clinical grounds, since various pathological mechanisms lead to DCM (3). Recently, several TFs that respond to developmental, mechanical, metabolic, and extracellular cues have been identified to be associated with regulating cardiac remodeling.
The CDK8 gene is composed of 13 exons and is located at chromosomal region 13q12.13. It encodes 464 amino acids to form a 53-kDa major isoform, which is an evolutionarily conserved serine/threonine kinase (8). CDK family members are known to play critical roles in different biological processes, such as metabolism, angiogenesis, spermatogenesis, neuronal differentiation, hematopoiesis, stem cell self-renewal, and carcinogenesis (21). As a special subtype of the CDK family, CDK8 has been suggested to act as both a positive and negative regulator of transcription (22).
CDK8 is a component of the Mediator complex, which is a large multi-subunit complex that plays a central role in the transcription regulation of eukaryotes. CDK8 and its regulatory subunits, MED12, MED13, and cyclin C, constitute the Mediator transcriptional regulatory complex. The Module is a 4-subunit complex that reversibly binds to the Mediator and regulates its activity by changing the structure and function of the core Mediator. The Mediator serves as a bridge in both transcriptional activation and repression by phosphorylation of the carboxy-terminal domain of the largest subunit of RNA polymerase II. The phosphorylation targets of CDK8 include the core Mediator-RNA-polymerase-II complex and many other transcriptional regulators, such as tissue-specific and general TFs, chromatin-remodeling factors, pause or release factors, and histone H3 (23).
Most previous research has focused on the role of human CDK8 in carcinogenesis as an oncogene, and its function to be a therapeutic target in many cancers (24). It has been demonstrated that the CDK8 gene is frequently amplified in colorectal cancer (CRC) (25,26), and overexpression of CDK8 has also been demonstrated in the pathogenesis of breast cancer, ovarian cancer (27), melanoma (28), acute myeloid leukemia (29), and prostate cancer (30). CDK8 modulates β-catenin-mediated transcription directly or indirectly. Knockdown of CDK8 induces aberrant activation of the Wnt/β-catenin signaling pathway in CRC. However, the role of CDK8 in cancer is context-dependent. For example, in contrast to the findings above, deletion of CDK8 in a murine intestinal tumor model induced increased tumor size and growth rate, which resulted in reduced survival (24). Although many studies have reported the successful development of effective CDK8 inhibitors, many of them have shown unacceptable toxicity (31). The toxicity of these CDK8 inhibitors might provide insights into the essential functions of CDK8 in the process of normal development (8). Several in vitro and in vivo experiments have demonstrated that deficiency of CDK8 causes developmental arrest of embryos through transcriptional deregulation of developmentally critical genes, resulting in embryonic lethality (32).
A recent study reported by Calpena et al. (8) firstly assessed 12 patients with 8 heterozygous nonsynonymous variants of CDK8. All predicted substitutions localized at the ATP-binding pocket of the kinase domain, which led to a decrease in the phosphorylation level of the CDK8 substrate, STAT1-Ser727. Six of the 12 subjects suffered congenital heart defects (CHDs), classified as left ventricular obstruction and conotruncal heart anomaly. They pointed out that the developmental disorder caused by missense mutations in CDK8 has overlapping phenotypes with symptoms related to mutations of other subunits of the Mediator kinase module, thus indicating that there might be overlap in pathogenic mechanisms. Uehara et al. (7) reported 2 cases of CDK8-associated disorder involving two de novo missense mutations also within the kinase domain of CDK8, which suggested that craniofacial and cardiac defects are critical components of the CDK8-related disorder. A kinase assay of human CDK8 showed decreased kinase activity. A zebra fish model was used for functional analysis, which showed that non-synonymous variants in the kinase domain of CDK8 act as hypomorphic alleles that can cause human craniofacial and heart defects. These data demonstrate the essential role of CDK8 in prenatal development and cardiovascular disease. Hall et al. (12) found that CDK8 protein expression levels increased in human failing heart explants and identified that the same heart failure phenotype was present in cardiac-specific CDK8-expressing mice. Myofilament gene isoforms not typically expressed in adult cardiomyocytes were observed to be enriched, while oxidative phosphorylation and fatty acid biosynthesis genes were decreased. Interestingly, the expression levels of candidate upstream TFs and MAPK signaling pathways are considered to regulate cardiomyocyte size remained relatively unaffected. Their research revealed that manipulating cardiac gene expression through increased CDK8 levels is detrimental to the heart by novel differential regulation of a transcriptional profile enriched in sarcomeric and metabolic gene expression pathways, which induces pathological remodeling and eccentric hypertrophy in heart failure. Moreover, the tension-based model related to CDK8 may be able to predict DCM since CDK8 transgenic cardiomyocytes can re-express fetal myofilament genes, compromise the sarcomeric structure, and introduce cardiac dilation.
Furthermore, several previous studies have demonstrated that CDK8 plays a significant role as a regulator in several signaling pathways, including WNT, TGFβ/BMP, STAT1, SHH, and NOTCH (10,11), most of which have been reported to be associated with the pathogenesis of DCM, especially TGFβ/BMP. The TGFβ family of cytokines are important regulators of cell cycle, differentiation, apoptosis, and inflammation (33). CDK8 enhances the transactivation potential and turnover of receptor-regulated Smad proteins in the TGFβ/BMP pathways (34). The abnormal expression of TGFβ may mediate myocardial injury through immunizing inflammation, and can cause the occurrence and development of DCM by signal pathway dysfunction. The apoptosis of cardiomyocytes plays a key role in the progression of DCM-induced heart failure, while the AP-1/Smad signaling pathway has been identified as the final common pathway of cardiac myocyte apoptosis. Moreover, Euler-Taimor and Heger (35) revealed that myocardial fibrosis and myocardial cell apoptosis are the consequences of negative regulation of inhibitory SMADs proteins, which are activated by the TGFβ family. CDKs have been proven to be vital regulators of cell cycle progression and RNA transcription. It has been shown that several genetic and epigenetic events cause universal overactivity of cell CDKs, and their inhibition can lead to both cell cycle arrest and apoptosis (36). Therefore, once the biological equilibrium of CDK8 activity has been deregulated, there might be modification of these developmental signaling networks related to the pathogenesis of DCM.
The A genotype of both rs17083838 and rs7992670 were found to be risk factors of DCM in our study. The rs17083838 polymorphism has been linked with pituitary adenoma (37), but the clinical significance of rs7992670 has not yet been examined. Although the 2 SNPs are located in the intron regions of CDK8, they may affect the RNA secondary structure, which may cause abnormal gene regulation and phenotypes (38). It has been confirmed that introns and noncoding RNAs serve as endogenous network control molecules, which allow direct communication between genes and multitasking in eukaryotic genomes (39). In addition, CDK8 plays a vital role in the process of RNA transcription. Taken together, the two SNPs may cause DCM through the deregulation of gene-gene communication rather than changing the encoded protein. According to the results of our survival analysis, the two SNPs found within the CDK8 gene might be related to the poor prognosis of DCM.
Nevertheless, the prognosis of DCM is associated with an LVEF <35%, right ventricular involvement and NYHA functional class III or IV. Furthermore, various factors can worsen the prognosis of DCM, such as functional mitral regurgitation, ventricular arrhythmia, myocardial fibrosis, dyssynchronous ventricular contraction, and enlargement of other chambers. Exogenous factors, such as enteroviruses, have also been related to poor prognosis (40). The treatments for DCM mainly focus on reducing symptoms of heart failure and improving cardiac function. Management can also affect the prognosis. All the patients in this study were treated under the guidance of clinical guidelines. The current survival analysis showed that the two SNPs were related to a worse prognosis. This result reveals that genotypic variations might also be prognostic factors of DCM.
Consequently, CDK8 is a possible biological candidate gene that plays a vital role in the pathogenesis of some cardiovascular diseases, including DCM. However, up to now, no study or analysis has mentioned which SNPs of CDK8 are associated with susceptibility to DCM. We firstly defined the correlation between CDK8 SNPs (rs17083838 and rs7992670) and DCM in a Chinese Han population. However, due to the small sample size, regional differences, and ethnic differences, this study still has some limitations. The main results were obtained based on a regression model that established a relationship between SNPs of CDK8 and DCM. In the absence of identifying specific pathophysiological links between genotypes and phenomena, this model cannot be recognized as adequate. Furthermore, various factors can affect the prognosis of DCM. Hence, the positive results do not necessarily mean that the pathogenic SNPs have been found. Further studies of the pathogenic mechanisms with stricter screening, larger samples sizes, and more races are indispensable.
Conclusions
Taken together, we firstly revealed that SNPs rs17083838 and rs7992670 of the CDK8 gene might be associated with the higher DCM risk and worse prognosis in the Chinese Han population. In China, the Han population is the largest ethnic group, which is representative. The CDK8 gene is currently a research hotspot. CDK8 gene polymorphisms might be candidate risk factors for DCM in Chinese. Therefore, larger sample sizes of CDK8 SNPs in multiple populations would be required to confirm our findings. More studies are also necessary to investigate the specific mechanistic link between CDK8 and DCM, which may potentially promote the development of novel genetic therapies that involving CDK8.
Acknowledgments
We would like to thank Dr. Bo Zhang for his help in polishing our paper. And we also would like to thank AME Editing Service for polishing our paper.
Funding: This work was supported by the National Natural Science Foundation of China [No. 82071735]; The R&D Program for International S&T Cooperation and the Exchanges of Chengdu [No. 2020-GH02-00068-HZ] and Sichuan Science and Technology Program [No. 2021YFS0333, No. 2019YFS0350 and No. 2020ZYD007].
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/cdt-21-323
Data Sharing Statement: Available at https://dx.doi.org/10.21037/cdt-21-323
Peer Review File: Available at https://dx.doi.org/10.21037/cdt-21-323
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/cdt-21-323). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the West China Hospital of Sichuan University ethics committee (No. 81670346) and individual informed consents for this retrospective analysis were obtained.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Givertz MM, Mann DL. Epidemiology and natural history of recovery of left ventricular function in recent onset dilated cardiomyopathies. Curr Heart Fail Rep 2013;10:321-30. [Crossref] [PubMed]
- Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet J Rare Dis 2006;1:27. [Crossref] [PubMed]
- McNally EM, Golbus JR, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest 2013;123:19-26. [Crossref] [PubMed]
- Knöll RH, Hoshijima M, Chien KR. Mechanisms of dilated cardiomyopathies. Drug Discovery Today: Disease Mechanisms 2004;1:31-6. [Crossref]
- Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2008;29:270-6. [Crossref] [PubMed]
- McNally EM, Mestroni L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ Res 2017;121:731-48. [Crossref] [PubMed]
- Uehara T, Abe K, Oginuma M, et al. Pathogenesis of CDK8-associated disorder: two patients with novel CDK8 variants and in vitro and in vivo functional analyses of the variants. Sci Rep 2020;10:17575. [Crossref] [PubMed]
- Calpena E, Hervieu A, Kaserer T, et al. De Novo Missense Substitutions in the Gene Encoding CDK8, a Regulator of the Mediator Complex, Cause a Syndromic Developmental Disorder. Am J Hum Genet 2019;104:709-20. [Crossref] [PubMed]
- Galbraith MD, Andrysik Z, Pandey A, et al. CDK8 Kinase Activity Promotes Glycolysis. Cell Rep 2017;21:1495-506. [Crossref] [PubMed]
- Philip S, Kumarasiri M, Teo T, et al. Cyclin-Dependent Kinase 8: A New Hope in Targeted Cancer Therapy? J Med Chem 2018;61:5073-92. [Crossref] [PubMed]
- Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 2013;48:575-608. [Crossref] [PubMed]
- Hall DD, Ponce JM, Chen B, et al. Ectopic expression of Cdk8 induces eccentric hypertrophy and heart failure. JCI Insight 2017;2:e92476 [Crossref] [PubMed]
- Richardson P, McKenna W, Bristow M, et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation 1996;93:841-2. [Crossref] [PubMed]
- Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 2006;113:1807-16. [Crossref] [PubMed]
- Iranpur M Esmailizadeh AK. Rapid extraction of high quality DNA from whole blood stored at 4 ◦C for long period. 2010. Available online: http://www.protocol-online.org/cgi-bin/prot/page.cgi?g=print_page/4175.html
- Vieux EF, Kwok PY, Miller RD. Primer design for PCR and sequencing in high-throughput analysis of SNPs. Biotechniques 2002;28-30, 32. [Crossref] [PubMed]
- Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform 2012;10:117-22. [Crossref] [PubMed]
- Solé X, Guinó E, Valls J, et al. SNPStats: a web tool for the analysis of association studies. Bioinformatics 2006;22:1928-9. [Crossref] [PubMed]
- Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97-8. [Crossref] [PubMed]
- Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res 2009;19:519-23. [Crossref] [PubMed]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013;140:3079-93. [Crossref] [PubMed]
- Nemet J, Jelicic B, Rubelj I, et al. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 2014;97:22-7. [Crossref] [PubMed]
- Jeronimo C, Robert F. Kin28 regulates the transient association of Mediator with core promoters. Nat Struct Mol Biol 2014;21:449-55. [Crossref] [PubMed]
- Xi M, Chen T, Wu C, et al. CDK8 as a therapeutic target for cancers and recent developments in discovery of CDK8 inhibitors. Eur J Med Chem 2019;164:77-91. [Crossref] [PubMed]
- Firestein R, Bass AJ, Kim SY, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008;455:547-51. [Crossref] [PubMed]
- Tsafrir D, Bacolod M, Selvanayagam Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res 2006;66:2129-37. [Crossref] [PubMed]
- Eirew P, Steif A, Khattra J, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015;518:422-6. [Crossref] [PubMed]
- Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 2010;468:1105-9. [Crossref] [PubMed]
- Pelish HE, Liau BB, Nitulescu II, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 2015;526:273-6. [Crossref] [PubMed]
- Brägelmann J, Klümper N, Offermann A, et al. Pan-Cancer Analysis of the Mediator Complex Transcriptome Identifies CDK19 and CDK8 as Therapeutic Targets in Advanced Prostate Cancer. Clin Cancer Res 2017;23:1829-40. [Crossref] [PubMed]
- Dale T, Clarke PA, Esdar C, et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat Chem Biol 2015;11:973-80. [Crossref] [PubMed]
- Westerling T, Kuuluvainen E, Mäkelä TP. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 2007;27:6177-82. [Crossref] [PubMed]
- Macias MJ, Martin-Malpartida P, Massagué J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem Sci 2015;40:296-308. [Crossref] [PubMed]
- Alarcón C, Zaromytidou AI, Xi Q, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 2009;139:757-69. [Crossref] [PubMed]
- Euler-Taimor G, Heger J. The complex pattern of SMAD signaling in the cardiovascular system. Cardiovasc Res 2006;69:15-25. [Crossref] [PubMed]
- Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol 2006;24:1770-83. [Crossref] [PubMed]
- Ye Z, Li Z, Wang Y, et al. Common variants at 10p12.31, 10q21.1 and 13q12.13 are associated with sporadic pituitary adenoma. Nat Genet 2015;47:793-7. [Crossref] [PubMed]
- Sabarinathan R, Tafer H, Seemann SE, et al. RNAsnp: efficient detection of local RNA secondary structure changes induced by SNPs. Hum Mutat 2013;34:546-56. [Crossref] [PubMed]
- Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 2001;18:1611-30. [Crossref] [PubMed]
- Schultheiss HP, Fairweather D, Caforio ALP, et al. Dilated cardiomyopathy. Nat Rev Dis Primers 2019;5:32. [Crossref] [PubMed]


