
MiR-216a accelerates proliferation and fibrogenesis via targeting PTEN and SMAD7 in human cardiac fibroblasts
Introduction
Chronic heart failure (CHF) is a highly prevalent, progressive, multi-system disease, characterized by persistent low ventricular pump and/or filling function. Despite of numerous etiologies, ventricular remodeling is the basis for the development of CHF, including the reactive changes of cardiac myocytes and cardiac fibrosis, and the latter is the common pathology of HF caused by many etiologies (1). Cardiac fibrosis not only destroys the physiological elastic fiber skeleton of the heart, resulting in increased cardiac stiffness, which may directly contribute to both systolic and diastolic dysfunction in nearly all types of cardiac injury, but also provides potential electrophysiological matrix for cardiac death caused by malignant arrhythmias, seriously affecting the outcome of chronic HF (2,3). Cardiac fibroblasts play an important role in normal cardiac function, as well as in the fibrosis process during the CHF. Recently, studies have shown that plenty of microRNAs (miRNAs) such as miR-98, miR-19a-3p/19b-3p and miR-122 could provoke in the human cardiac fibroblasts (HCF) cells via activation of the TGF-β1 signaling pathways (4-6).
Exactly, miRNAs as a class of small noncoding RNAs binding incomplete complementary sequences of mRNAs, repressing translation or degrading target genes, have been found to be involved in the development of many disorders including HF (7,8). The alterations in their expression level might play crucial role in the ventricular remodeling, especially in the occurrence and progress of the cardiac fibrosis. The balancing and tight regulation of a specific miRNA might awaken the HCF cells and could be a key to drive the fibrogenesis (7,8). In this study, we showed that miR-216a was significantly up-regulated in the HF and over-expression of miR-216a promoted proliferation and enhanced the fibrogenesis in the HCF cells. Moreover, phosphatase and tensin homolog (PTEN) and mothers against decapentaplegic homolog 7 (SMAD7) were both the direct target genes of miR-216a, which were inhibited by miR-216a leading to the activation of Akt/mTOR and TGF-βRI/Smad2 in the HCF cells. Finally, we demonstrated here that miR-216a might regulate cardiac fibrosis, at least partially, via targeting the PTEN and SMAD7 in the HCF cells.
Methods
Participants and samples
Approval for this study was obtained from the medical ethics committee of the First Affiliated Hospital of Nanjing Medical University (The number of ethics is 2015-SRFA-085). Sixty-four patients with CHF caused by dilated cardiomyopathy (DCM) and ischemic cardiomyopathy (ICM) and 30 healthy controls (HC) were recruited in this study. Among the HF group, 34 and 30 patients were diagnosed as DCM and ICM, respectively. Written informed consent was obtained from all study participants. Inclusion criteria for the HF group were chronic stable HF diagnosed according to Framingham standards, New York Heart Association stage III–IV, and plasma prohormone of brain natriuretic protein (pro-BNP) content ≥1,000 ng/L. Exclusion criteria were defined as those patients with a history of myocardial infarction, percutaneous coronary intervention, coronary artery bypass grafting, coronary disease, iodine indolence, poor renal function, or hemodynamically unstable patients.
The characteristics of the HF and control groups were shown in Table 1. The numbers of patients taking diuretics (64 patients), beta-adrenoceptor antagonists (62 patients), renin-angiotensin-aldosterone system (RAAS) inhibitors (60 patients), aspirin (30 patients), and statins (28 patients) were recorded. 15 patients had a history of arrhythmia (paroxysmal atrial fibrillation and/or nonsustained ventricular tachycardia), and 8 of them had implantable cardiac electrical defibrillators.

Full table
Plasma isolation and storage
Whole venous blood sample was drawn from each participant. Samples were initially collected in ethylenediaminetetraacetic acid (EDTA)-containing tubes (Becton, Dickinson and Company) followed by a two-step centrifugal process (350 RCF (reactive centrifugal force) for 10 min and 20,000 RCF for 10 min (Beckman Coulter, USA) to isolate cell-free plasma samples within 12 hours. The obtained plasma samples were then restored in RNase-free tube at −80 °C ready for future analysis. All of the collected plasma avoided freeze-thaw cycles.
miRNAs profiling screening and pathway enrichment analysis of differentially expressed genes (DEGs) in HF
Expression profiling of miRNAs and mRNA by array or high throughput sequencing of human HF specimen were searched in Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Two datasets of GSE53080 and GSE57338 were collected for further analysis because of larger sample size and relatively higher reliability of data.
The GSE53080 data derived from high throughput sequencing of myocardial and circulating miRNAs in human HF was downloaded and the “edgeR” (Empirical Analysis of Digital Gene Expression Data in R) was used to screen the miRNAs profiling between non-failing and failing heart samples. The significance analysis with |fold change (FC)| >2 and adj.P value <0.01 were chosen as the cut-off criteria to select miRNAs as the miRNA characteristic profiling of HF. The common variation of miRNAs in the myocardial and plasma samples of HF caused by DCM and ICM were collected for next step verification.
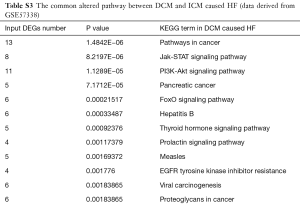
The GSE57338 data identified the myocardial gene expression signatures of HF by microarray. The GEO2R tool was used to find the DEGs between the non-failing and failing heart samples caused by DCM and ICM, respectively. The DEGs were then uploaded to the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/), an online program providing a comprehensive set of functional annotation tools for investigators to understand biological meaning behind large list of genes, to perform KEGG pathway enrichment analysis. Those common terms including DEGs in the myocardial of HF caused by DCM and ICM were selected as the key biological process and pathways. P<0.05 was set as the cut-off criterion.
Plasma miRNAs extraction
Total miRNAs were extracted using the mirVana PARIS Kit (Ambion, Austin, TX, USA) for each 200 µL plasma sample following the manufacturer’s instructions. The acquired total miRNAs were lysed into 100 µL RNase-free water and kept at −80 °C until analysis. The ultraviolet spectrophotometer was applied to evaluate the concentration and purity of total miRNAs samples. If the concentration of total miRNAs was less than 10 ng/µL, it was not included in data analysis. During the process, additional 5 µL of synthetic C.elegans miR-39 (cel-miR-39) (5 nM/L, RiboBio, Guangzhou, China) was added to each sample after denaturing solution (Ambion, Austin, TX, USA) for sample-to-sample normalization.
qRT-PCR for miR-216a detection
miRNAs were amplified using Bulge-LoopTM miRNA quantitative reverse transcription polymerase chain reaction (qRT-PCR) Primer Set (RiboBio, Guangzhou, China) with specific primers of RT and PCR. According to previous study, RT and PCR procedures were performed on 7900HT real-time PCR system (Applied Biosystems, Foster City, CA, USA) in the condition of 42 °C for 60 min followed by 70 °C for 10 min (for RT) and 95 °C for 20 sec, followed by 40 cycles of 95 °C for 10 sec, 60 °C for 20 sec and then 70 °C for 10 sec (for PCR), respectively. SYBR Green (SYBR® Premix Ex TaqTM II, TaKaRa, Dalian, China) was used to calculate the amount of PCR products by the level of fluorescence and melting analysis was introduced to evaluate the specificity of PCR products. As our previously described, the expression levels of plasma miR-216a relative to exogenous reference miRNA (cel-miR-39) were quantified by the value of 2−ΔCt (ΔCt = Ct miR-216a − Ct cel-miR-39) and then the relative FCs of plasma miR-216a between HF and HC groups were calculated by the value of 2−ΔΔCt (ΔΔCt = the average ΔCt of HF group − the average ΔCt of HC group) (5).
Cell culture and treatment
The HCF cell line was purchased from Cell Bank of Tongpai Biotechnology Co., Ltd. (Shanghai, China). Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco Inc.) was formulated as the cell culture medium for HCF cells. In order to make the complete growth medium, the following components of 4.5 g/L glucose and fetal bovine serum were added in the base medium with the final concentration of 10%. Exponentially growing cultures were maintained in a humidified atmosphere of 5% carbon dioxide (CO2) at 37 °C.
Transient transfections
HCF cells were seeded in 6-well plates (6×105 cells/well). HCF cells were transfected with miR-216a mimic (100 nM) or miRNA mimic control (100 nM) using LipofectamineTM 2000 reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol, respectively. The miR-216a mimic and miRNA mimic control were purchased from Integrated Biotech Solutions Co., Ltd (Shanghai, China).
Cell proliferation assay
The cell counting kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc., Japan) method was used to analyze the cell growth of HCF cells according to the manufacturer’s instructions. Six hours after the transfection of miR-216a mimic or miRNA mimic control in the HCF cells, the cells were then seeded with a density of 5×103 cells/well in 96-well plates for total 24, 48, 72 hours at 37 °C with 5% CO2, respectively. The medium of each well was then substituted with 100 µL of fresh medium containing 10% CCK-8, and the cultures were incubated at 37 °C for 2 hours. The absorbance value (A) was determined using SynergyTM 2 Multi-Mode Microplate Reader (BioTek Instruments, Inc., headquartered in Winooski, VT, USA) at 450 nm. Cell viability (%) = average absorbance of treated group/average absorbance of control group ×100%.
Clonogenic assay
HCF cells were transfected with miR-216a mimic or miRNA mimic control and then plated in 6-well plates at a density of 250 cells per well, incubated at 37 °C for two weeks, fixed, and stained with crystal violet. Colonies containing more than 50 cells were counted under a microscope from three independent replicates. Results are expressed as mean ± SD.
Dual-luciferase activity assay
The 3’UTRs of human PTEN and SMAD7 cDNA containing the putative target site for the miR-216a (sequence shown in Supplementary data) were chemically synthesized and inserted at the XbaI site, immediately downstream of the luciferase gene in the pGL3-control vector (Promega, Madison, WI, USA) by Integrated Biotech Solutions Co., Ltd (Shanghai, China), respectively. Twenty-four hours before transfection, cells were plated at 1.5×105 cells/well in 24-well plates. Two hundred ng of pGL3-PTEN-3’-UTR or pGL3-SMAD7-3’-UTR plus 80 ng pRL-TK (Promega) were transfected in combination with 50 nM of the miR-216a mimic or miRNA mimic control using LipofectamineTM 2000 reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol, respectively. Luciferase activity was measured 24 hours after transfection using the Dual Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to renilla luciferase activity for each transfected well. Three independent experiments were performed in triplicate.
Western blotting
HCF cells were seeded in 6-well plates (6×105 cells/well), cultured for 72 hours followed by transfection with miR-216a mimic or miRNA mimic control. Total protein was extracted from the cells and separated on 10% sodium dodecyl sulfate-polyacrylamide gels. Western blot was carried out as our previously described (5). Antibodies for the fibrogenesis indicator proteins of collagen I α2 (catalog: ab96723) and fibronectin (catalog: ab32419); antibodies for the miR-216a targets of Smad7 (catalog: ab90086) and PTEN (catalog: ab32199); antibodies for effector proteins of PI3K-Akt and TGF-β1-Smads signal pathways including phosphorylated-pan-Akt (catalog: ab38449)/phosphorylated-mTOR (catalog: ab1093) and TGF-βRI (catalog: ab31013)/phosphorylated-Smad2 (catalog: ab53100) and antibodies for reference protein of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (catalog: ab8245) were all purchased from Abcam. The expression level of each protein was normalized to that of GAPDH and FCs were calculated.
Statistical analysis
All experiments were carried out more than 3 times. Numerical results are presented as mean ± SD. Student’s t-test was used to analyze the difference between means using SPSS13.0 software (Chicago, IL). Significance was indicated as *P<0.05, #P<0.05, **P<0.01, ##P<0.01 and ***P<0.001. * and # represented different gene in the same figure.
Results
MiR-216a was closely related to HF
The miRNAs profiling screening of HF was based on the high throughput sequencing data from the dataset of GSE53080. The miRNAs signatures in the left-ventricular myocardium and the plasma of HF patients caused either by DCM or ICM were shown in Tables S1,S2, respectively. Analysis of the common miRNAs alterations in both the myocardium tissues and the plasma of HF patients caused either by DCM or ICM verified that the mature members of miR-216a and miR-217 derived from the miR-216a/217 cluster were significantly up-regulated (Figure 1A), especially the miR-216a, while no miRNAs with obvious down-regulated expression level in common was found (Figure 1B).

Full table
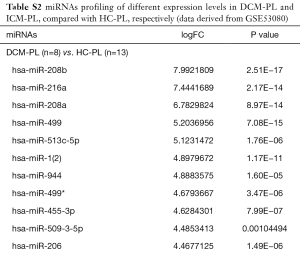
Full table
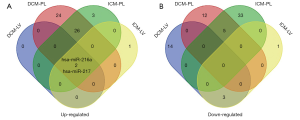
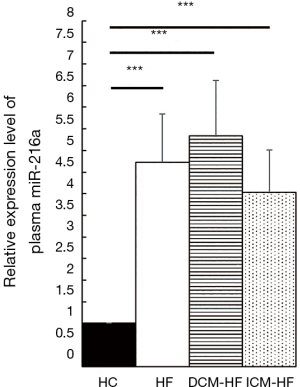
MiR-216a/217 cluster was on the chr2 and could be detected in endothelial cells exposed to hypoxia via deep-sequencing (9). As the pathophysiological essence of CHF was long-term hypoxia of organs (10), the mature members of the miR-216a/217 cluster might be also involved in HF. Exactly, the qRT-PCR further confirmed that the miR-216a was significantly up-regulated in the plasma of the HF patients with the average fold increment of 4.727, compared with the HCs (Figure 2, ***P<0.001). Moreover, the plasma miR-216a was consistently over-expressed in HF patients caused either by DCM or ICM, with the average FCs of 5.336 and 4.036, respectively (Figure 2, ***P<0.001). Those suggested that miR-216a was closely related to HF.
MiR-216a promoted proliferation and enhanced the fibrogenesis in HCF cells
Since the fibrosis served as the pathological matrix of the HF, the potential role of miR-216a on HCF cells was studied. In HCF cells, the CCK-8 cell proliferation assay revealed that cells transfected with miR-216a mimic exhibited a significant increase of cell viability, compared with those transfected with miRNA mimic control. The P value of 24 hours, 48 hours and 72 hours are 0.037, 0.005, 0.002, respectively (Figure 3A). Clonogenic assay also confirmed that miR-216a could accelerate the proliferation of HCF cells. Those transfected with miR-216a mimic showed strong enhancement of clonogenic ability, compared with those transfected with miRNA mimic control (Figure 3B, P=0.003). Meanwhile, the indicator proteins of the fibrogenesis of collagen I α 2 and fibronectin were significantly up-regulated in those HCF cells transfected with miR-216a mimic, compared with those transfected with miRNA mimic control (Figure 3C, **P=0.007, ##P=0.005). The above indicated that miR-216a could promote the proliferation and enhance the fibrogenesis in HCF cells, which might induce the fibrosis in HF.

PTEN and SMAD7 were the direct target genes of miR-216a
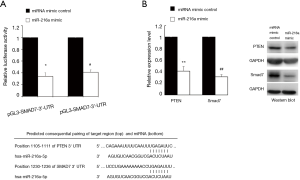
TargetScan predicted that both PTEN and SMAD7 were targets of miR-216a in various species. To verify whether these two genes were indeed the targets of miR-216a, we carried out luciferase reporter assays using vectors harboring the 3’UTR of PTEN or SMAD7 with the putative target site for miR-216a downstream of the luciferase gene (pGL3-PTEN-3’-UTR and pGL3-SMAD7-3’-UTR), respectively. HCF cells were transfected with luciferase reporter vectors and miR-216a mimic or miRNA mimic control, respectively. In HCF cells, the relative luciferase activity was found to be significantly decreased when pGL3-PTEN-3’-UTR or pGL3-SMAD7-3’-UTR was transfected together with miR-216a mimic but not with the miRNA mimic control. These results confirmed that both PTEN and SMAD7 were targets of miR-216a (Figure 4A, *P=0.029, #P=0.044). Moreover, 72 hours after the transfection in HCF cells, western blot revealed that the protein expression levels of PTEN and SMAD7 were significantly down-regulated in cells transfected with miR-216a mimic relative to those transfected with miRNA mimic control (Figure 4B, **P=0.006, ##P=0.005). All these results confirmed that both PTEN and SMAD7 were the direct targets of miR-216a.
Akt/mTOR and TGF-βRI/Smad2 were activated by miR-216a in HCF cells
The common pathway enrichment analysis of the DEGs in HF caused by DCM and ICM was shown in Table S3. Among the common altered signal pathways between the HF caused by DCM and ICM, the PI3K-Akt signal pathway was with the most significance, which suggested that this signal pathway was deeply involved in the pathological molecular process of the HF. Moreover, plenty of studies including of our previous study had elucidated that the TGF-β1-Smads signal pathway plays an important role in the cardiac fibrosis via enhancing the fibrogenesis in HCF cells (4-6).
Full table
It is well known that the PTEN antagonizes the PI3K-Akt signaling pathway while Smad7 is the antagonist of TGF-β1-Smads signaling. Down-regulation of PTEN causes the activation of Akt which phosphorylates the mTOR, leading to the cell proliferation (11), while inhibition of Smad7 which targets the TGF-β1 receptors especially the TGF-βRI for degradation through ubiquitination (12), promotes the TGF-β1-Smads signaling. Since both PTEN and SMAD7 were the direct targets of miR-216a as mentioned above, we hypothesized that the activation of Akt/mTOR and TGF-βRI/Smad2 in HCF cells could be induced by miR-216a, which constituted the intrinsic molecular mechanism of proliferation promoting and fibrogenesis enhancement by miR-216a in HCF cells.
To verify this, HCF cells were transfected with miR-216a mimic and miRNA mimic control to detect changes in the expression levels of the effector proteins of P-Akt/P-mTOR and TGF-βRI/P-Smad2, which belonged to PI3K-Akt and TGF-β1-Smads signal pathways, respectively. In HCF cells, at 72 hours after transfection, western blot revealed that the expression of P-Akt, P-mTOR, TGF-βRI and P-Smad2 were all significantly up-regulated in cells transfected with miR-216a mimic compared with those transfected with miRNA mimic control, respectively (Figure 5A, **P=0.006, ##P=0.005; Figure 5B, **P=0.004, ##P=0.003).
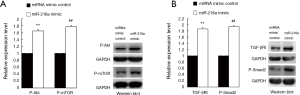
Taken all together, we found that miR-216a accelerates proliferation and fibrogenesis in HCF cells via targeting PTEN and SMAD7 leading to the activation of Akt/mTOR and TGF-βRI/Smad2.
Discussion
CHF is a progressive disease with relatively poor prognosis and lacks effective therapy. Myocardial fibrosis leading to ventricular remodeling is a common pathology of CHF, although there are many factors (1-3). Aberrant expressions of miRNAs act as biomarkers in the development and progression of CHF. MiRNAs are widely involved in the process of CHF, including interstitial fibrosis, myocardial hypertrophy, myocardial ischemia, and cellular proliferation and apoptosis (7). In this study, analysis of the common miRNAs alterations was performed on both the myocardium tissues and the plasma of HF patients and we identified 2 up-regulated miRNAs. The expression of miR-216a was then confirmed markedly increased in the plasma of HF patients. Previous studies have described the role of miR-216a in HF. Menghini et al. found that mir-216a had a certain effect on the pathogenesis of cardiovascular disorders and atherosclerosis by controlling ox-LDL induced autophagy in HUVECs (13). Barsanti showed that miR-216a-5p was involved in focal adhesion/integrin pathway and in actin cytoskeleton regulation (14). However, very little was known about miR-216a and its biological effects on myocardial fibrosis. In our study, miR-216a was explored in our study and over-expression of miR-216a significantly was founded to promote viability, proliferation and enhanced the fibrogenesis in HCF cells. Taken together, these results indicate that miR-216a acts as a pathogenetic miRNA exerting an important effect on CHF progression.
PTEN is localized in chromosome 10, a lipid phosphatase and dual specificity protein. The lipid phosphatase activity is critical for its suppressor function, which negatively regulate PI3K-Akt signaling pathway by dephosphorylation and thereby modulate cell cycle progression and cell survival (11). Deleting PTEN was recently revealed to have a positive effect on cell proliferation and neuronal growth during development (15). Zhou et al. reported that downregulating expression of PTEN and Smad7 activate profibrotic signaling pathways (16). SMAD7 has been identified as a member of SMAD family, binding SMURF2 and targeting the TGF-β1 receptors especially the TGF-βRI for degradation through ubiquitination (12).
It has been reported that degradation of Smad7 may serve as a key mechanism for AF-induced atrial fibrosis (17). Lei et al. found that increasing Smad7 expression results in reduced collagen production and α-smooth muscle actin expression in the activated HSCs (18). Those indicated that both PTEN and SMAD7 were closely related with fibrosis. In this study, we found that PTEN and SMAD7 were the direct targets of miR-216a in CHF. MiR-216a down-regulated PTEN and SMAD7 expression post-transcriptionally through binding to the 3’UTR of PTEN and SMAD7 which was verified by the luciferase reporter assay and western blot analysis.
In addition, our data also suggested that miR-216a/PTEN axis regulated cell viability and cell proliferation by Akt/mTOR signaling pathways and miR-216a/SMAD7 axis regulated fibrogenesis by involving in the TGF-βRI/Smad2 signaling pathways. It has been well known that Akt/mTOR signaling pathway has an effect on cell cycle and proliferation and TGF-βRI/Smad2 signaling pathway plays a key role in cell fibrogenesis (19,20). Since PTEN is a negative regulator of Akt/mTOR signaling pathways and SMAD7 antagonize of TGF-β1-Smads signaling (11,12), our study further showed that miR-216a mimic heighten the activation of the above two signaling pathways in the HCF cells. Therefore, we considered that miR-216a activated the above two signaling pathways by direct degradation of PTEN and SMAD7 in HCF cells. However, the specific mechanisms underlying the activation remains poorly understood and further investigations are needed.
Conclusions
In conclusion, the results of our study showed that miR-216a was up-regulated in both tissues and the plasma of HF patients. It might be a potential marker for early HF screening. Importantly, miR-216a might act as a promoter of cardiac fibrosis through inducing proliferation and enhancing fibrogenesis in the HCF cells, at least via direct targeting the PTEN and SMAD7, leading to the activation of Akt/mTOR and TGF-βRI/Smad2 signals in the HCF cells. These data might provide a promising approach for the treatment of CHF in the future.
Supplementary
The sequence of 3'UTR of human PTEN cDNA containing the putative target site for hsa-miR-216a, sequence in bold stands for the putative target site:
TAACGACTTCTCCATCTCCTGTGTAATCAAGGCCAGTGCTAAAATTCAGATGCTGTTAGTACCTACATCAGTCAACAACTTACACTTATTTTACTAGTTTTCAATCATAATACCTGCTGTGGATGCTTCATGTGCTGCCTGCAAGCTTCTTTTTTCTCATTAAATATAAAATATTTTGTAATGCTGCACAGAAATTTTCAATTTGAGATTCTACAGTAAGCGTTTTTTTTCTTTGAAGATTTATGATGCACTTATTCAATAGCTGTCAGCCGTTCCACCCTTTTGACCTTACACATTCTATTACAATGAATTTTGCAGTTTTGCACATTTTTTAAATGTCATTAACTGTTAGGGAATTTTACTTGAATACTGAATACATATAATGTTTATATTAAAAAGGACATTTGTGTTAAAAAGGAAATTAGAGTTGCAGTAAACTTTCAATGCTGCACA.
The sequence of 3'UTR of human SMAD7 cDNA containing the putative target site for hsa-miR-216a, sequence in bold stands for the putative target site:
ATACACTCGTATGATACTTCGACACTGTTCTTAGCTCAATGAGCATGTTTAGACTTTAACATAAGCTATTTTTCTAACTACAAAGGTTTAAATGAACAAGAGAAGCATTCTCATTGGAAATTTAGCATTGTAGTGCTTTGAGAGAGAAAGGACTCCTGAAAAAAAACCTGAGATTTATTAAAGAAAAAAATGTATTTTATGTTATATATAAATATATTATTACTTGTAAATATAAAGACGTTTTATAAGCATCATTATTTATGTATTGTGCAATGTGTATAAACAAG
Acknowledgments
Funding: This study was funded by the National Natural Science Foundation of China (grant No. 81571873), Jiangsu Province “333” project (grant No. BRA2017547) and Jiangsu Province Key Medical Talents project (grant No. ZDRCA2016016).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animals in this study were obtained from the Animal Experimental Center of Nanjing Medical University. All animals handling and procedures were approved by the Animal Ethics Committee of Nanjing Medical University.
References
- Segura AM, Frazier OH, Buja LM, et al. Fibrosis and heart failure. Heart Fail Rev 2014;19:173-85. [Crossref] [PubMed]
- Passino C, Barison A, Vergaro G, et al. Markers of fibrosis, inflammation, and remodeling pathways in heart failure. Clin Chim Acta 2015;443:29-38. [Crossref] [PubMed]
- Schirone L, Forte M, Palmerio S, et al. A Review of the Molecular Mechanisms Underlying the Development and Progression of Cardiac Remodeling. Oxid Med Cell Longev 2017;2017:3920195. [Crossref] [PubMed]
- Cheng R, Dang R, Zhou Y, et al. MicroRNA-98 inhibits TGF-β1-induced differentiation and collagen production of cardiac fibroblasts by targeting TGFBR1. Human Cell 2017;30:192-200. [Crossref] [PubMed]
- Zou M, Wang F, Gao R, et al. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced fibrogenesis in human cardiac fibroblasts. Sci Rep 2016;6:24747. [Crossref] [PubMed]
- Beaumont J, López B, Hermida N, et al. microRNA-122 down-regulation may play a role in severe myocardial fibrosis in human aortic stenosis through TGF-β1 up-regulation. Clin Sci (Lond) 2014;126:497-506. [Crossref] [PubMed]
- Chen C, Ponnusamy M, Liu C, et al. MicroRNA as a Therapeutic Target in Cardiac Remodeling. Biomed Res Int 2017;2017:1278436. [Crossref] [PubMed]
- Travers JG, Kamal FA, Robbins J, et al. Cardiac Fibrosis: The Fibroblast Awakens. Circ Res 2016;118:1021-40. [Crossref] [PubMed]
- Voellenkle C, Rooij JV, Guffanti A, et al. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA 2012;18:472-84. [Crossref] [PubMed]
- Zhao DS, Chen Y, Jiang H, et al. Serum miR-210 and miR-30a expressions tend to revert to fetal levels in Chinese adult patients with chronic heart failure. Cardiovasc Pathol 2013;22:444-50. [Crossref] [PubMed]
- Costa HA, Leitner MG, Sos ML, et al. Discovery and functional characterization of a neomorphic PTEN mutation. Proc Natl Acad Sci U S A 2015;112:13976-81. [Crossref] [PubMed]
- Kavsak P, Rasmussen RK, Causing CG, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 2000;6:1365-75. [Crossref] [PubMed]
- Menghini R, Casagrande V, Marino A, et al. MiR-216a: a link between endothelial dysfunction and autophagy. Cell Death Dis 2014;5:e1029. [Crossref] [PubMed]
- Barsanti C, Trivella MG, D'Aurizio R, et al. Differential regulation of microRNAs in end-stage failing hearts is associated with left ventricular assist device unloading. Biomed Res Int 2015;2015:592512. [Crossref] [PubMed]
- Knafo S, Esteban JA. PTEN: Local and Global Modulation of Neuronal Function in Health and Disease. Trends Neurosci 2017;40:83-91. [Crossref] [PubMed]
- Zhou X, Zang X, Ponnusamy M, et al. Enhancer of Zeste Homolog 2 Inhibition Attenuates Renal Fibrosis by Maintaining Smad7 and Phosphatase and Tensin Homolog Expression. J Am Soc Nephrol 2016;27:2092-108. [Crossref] [PubMed]
- He X, Gao X, Peng L, et al. Atrial fibrillation induces myocardial fibrosis through angiotensin II type 1 receptor-specific Arkadia-mediated downregulation of Smad7. Circ Res 2011;108:164-75. [Crossref] [PubMed]
- Lei XF, Fu W, Kim-Kaneyama JR, et al. Hic-5 deficiency attenuates the activation of hepatic stellate cells and liver fibrosis through upregulation of Smad7 in mice. J Hepatol 2016;64:110-7. [Crossref] [PubMed]
- Yang W, Wu Z, Yang K, et al. BMI1 promotes cardiac fibrosis in ischemia-induced heart failure via the PTEN-PI3K/Akt-mTOR signaling pathway. Am J Physiol Heart Circ Physiol 2019;316:H61-9. [Crossref] [PubMed]
- Okuno K, Naito Y, Yasumura S, et al. Influence of dietary iron intake estriction on the development of hypertension in weanling prehypertensive rats. Heart Vessels 2018;33:820-5. [Crossref] [PubMed]


