
Hypermethylation of miR-181b in monocytes is associated with coronary artery disease and promotes M1 polarized phenotype via PIAS1-KLF4 axis
Introduction
Arteriosclerotic cardiovascular disease (ASCVD) is the leading cause of morbidity and mortality worldwide (1). Atherosclerosis is considered as a chronic inflammatory and multifactorial disease. Endothelial activation and recruitment, vascular smooth muscle cell (VSMC) proliferation and migration, monocyte infiltration, macrophage polarization and foam cell formation are involved in atherogenesis. In this respect, macrophage played pivotal roles in all stages of atherosclerosis, from lesion initiation, expansion and rupture (2). Apart from a minority of macrophage-like cells derived from smooth muscle cells, resident macrophages in plaques are primarily from circulating monocytes (3). Furthermore, resident macrophages switch to different phenotypes in response to diverse environments and intracellular signaling pathways during the different phases of atherosclerosis (4). The different phenotypes of macrophages allow them to evolve to homeostatic imbalance in intracellular lipids, metabolites and inflammatory response. In this regard, activated macrophages are capable of polarizing towards a classical phenotype M1 induced by interferon-γ (IFN-γ) and the broad-spectrum Toll-like receptor (TLR) ligand lipopolysaccharide (LPS). An alternative phenotype M2 is induced by T helper 2 (Th2) cytokines interleukin-4 (IL-4) (5,6). Recent researches propose a series combination of markers to ascertain and characterize macrophage polarization (7,8). There is also extensive evidence suggesting that depletion of KLF4 in macrophages and vascular smooth muscle cells (VSMCs) resulted in remarkable reductions in plaque burden and increases in plaque stability (9). In addition, modulation of macrophage and VSMC phenotype switch and inflammation via KLF4 plays a critical role in atherogenesis (10,11).
MiRNAs are considered as small non-coding RNAs that play an important role in the post-transcriptional regulation of gene expression by blocking mRNA translation or inducing mRNA degradation. Accumulating epidemiological trials and experimental studies have demonstrated that miRNAs are involved in the development of atherosclerosis (12-14). More specifically, RNA sequencing followed with quantitative RT-PCR has obtained differentiated miRNA expression profiles between M1 and M2 polarization and determined the involvement of miR-181b, miR-34a, miR-223 and miR-125b in macrophage activation and polarization (12). DNA methylation, another important epigenetic pattern, occurs when methyl groups are added to cytosines by DNA methyltransferases (DNMTs). The methylated cytosines primarily reside in CpG islands around transcription start sites and mitigate gene transcription by blocking binding sites for transcription factors. Growing bodies of evidence suggest that aberrant DNA methylation in specific cell types emerge as promising biomarkers for cardiovascular diseases (15). Likewise, DNA methylation play causal roles in miRNA dysregulation and chronic inflammation in macrophages (16). DNA methylation also participates in the initiation and progression of atherosclerosis (15). However, the crosstalk between DNA methylation and miRNAs driving macrophage polarization and atherogenesis remains poorly understood.
In this study, we sought to explore the expression levels and methylation levels of candidate miRNAs in monocytes from healthy controls and patients with coronary artery disease (CAD). Furthermore, we determined the methylation status of candidate miRNA during macrophage polarization. Cellular and molecular experiments were also performed to explore the mechanism underlying candidate miRNA and macrophage polarization.
Methods
Study population
The clinical trial was approved by the First Affiliated Hospital of Harbin Medical University Ethics Committee (Ethical approval ID: HMU-2018-124). The informed written consent was provided in accordance with the Declaration of Helsinki (as revised in 2013) and obtained from the whole enrollment.
A total of 150 patients who were suspicious with presentations and symptoms related to CAD received angiography as routine diagnostic procedure. Patients who were complicated with congenital heart disease, cardiomyopathy, autoimmune diseases, acute infections, chronic obstructive pulmonary disease (COPD), tuberculosis, severe kidney or liver diseases, and cancers were excluded from the study.
Quantification of coronary plaques
At least six projections of left and right coronary artery were obtained using coronary angiography in each individual. The segments of coronary plaques were assessed according to the 18-segment SCAI classification (17). In brief, significant CAD was defined as the presence of luminal diameter narrowing ≥50% in the left anterior descending artery, left circumflex artery, right coronary artery and their main branches. Left main trunk stenosis was considered as two-vessel disease. The burdens of atherosclerotic plaques were evaluated by the length of plaque (>20 mm as long lesions), the degree of stenosis with and without calcification, and the number of coronary vessels with significant stenosis. The experts recorded the highest degree of stenosis in CAD patients with multivessel diseases for further investigation.
Monocyte isolation and purification
Leukocyte subsets separation and purification were performed immediately after the blood draw as previously described with minor modification (18). Briefly, peripheral blood mononuclear cells (PBMCs) and granulocytes were separated using density centrifugation on Ficoll-Paque (Sigma, USA), followed by centrifugation at 350 g for 20 minutes. Monocytes were purified from PBMCs by positive selection using anti-CD14 antibodies conjugated paramagnetic microbeads (Miltenyi Biotec, Germany), then the cells were separated according to standard protocols for magnetic-activated cell sorting (MACS) using LS columns and MidiMACS separator (Miltenyi Biotec, Germany). A separate sample of 1×105 monocytes was taken from individual for purity validation by flow cytometer (BD Biosciences, USA).
MiRNA mimics and inhibitor transfection
MiR-181b mimics, miR-181b inhibitors (anti181b), small interfering against PIAS1 (siPIAS) and the corresponding negative control (NC) were purchased from Genepharma Incorporation (Genepharma, China). The purified cells were transfected with miR-181b mimics, miR-181b inhibitors, siPIAS1 or their corresponding NC using Hiperfect transfection reagent (Qiagen Incorporation, Germany) based on the manufacturer’s instructions.
Induction of macrophage polarization
Following apheresis, circulating monocytes were plated (24 well plates, 3×105 cells per well), allowed to adhere for 24 hours, washed with culture solutions and cultured in 20% FBS/RPMI with recombinant M-CSF at a concentration of 100 ng/mL (Peprotech Incorporation, USA) for 4 days (M0 phenotype). Macrophages were washed and treated for 12 hours with RPMI +5% FBS alone, LPS at a concentration of 100 ng/mL and recombinant interferon gamma (IFN-γ, Peprotech Incorporation, USA) at a concentration of 20 ng/mL for differentiation to the M1 phenotype, or recombinant IL-4 at a concentration of 20 ng/mL (Peprotech Incorporation, USA) for differentiation to the M2 phenotype. To determine whether STAT1 mediated the role of miR-181b in macrophage polarization, macrophages were pretreated with 50 µM of Fludarabine (Selleck, USA) for 12 hours.
DNA extraction, bisulfite conversion and MethyLight PCR
Briefly, the genomic DNA was isolated by using DNeasy blood and tissue kit (Qiagen, Germany). All unmethylated cytosines in genomic DNA were converted to uracil by sodium bisulfite conversion (EpiTect kit, Qiagen, Germany), while the methylated cytosines were protected. MethyLight PCR was used to determine DNA methylation at the promoter of miR-181b as previously described (19). The percentage methylated of reference (PMR) value was calculated. CpGenome Universal Methylated DNA (Thermo Fisher, USA) served as a positive control. The exact methylation levels of miR-181b measured by methylated PCR were presented by PMR value relative to the positive control as described previously (19). The primers designed for MethyLight PCR are listed in Table S1.
Full table
Pyrosequencing quantitative methylation analysis
From selected CpG island, a DNA fragment containing 4 CpG dinucleotides was PCR-amplified (Table S1). The PCR products were subjected to pyrosequencing on PyroMark Q24 (QIAGEN, Valencia, CA, USA). We calculated the methylation index of each CpG dinucleotide by the PyroMark Q24 analysis software.
Dual luciferase reporter assay
HEK293T cells were cultured in DMEM (Gibco, USA) containing 10% FBS (Gibco, USA), 1% penicillin and streptomycin. PIAS1 3'UTR was cloned into the dual-luciferase reporter plasmid psiCHECK-2 (Promega, USA). Constructs carrying the fragment of the mutated PIAS1 3' untranslated coding regions (UTR) without the putative miR-181b binding sequence served as the mutated control. HEK293T cells were co-transfected with psiCHECK-PIAS1-3'UTR (Luci-PIAS1-WT) or with psiCHECK-PIAS1-3'UTR mutant (Luci-PIAS1-Mut), and miR-181b mimics using Lipofectamine 2000 for 24 hours. Furthermore, pGL4.45 (luc2P/ISRE/Hygro) plasmid was purchased from Promega corporation (Promega, USA) and was used to determine the transcription activity of STAT1 responsive elements.
Specific primers containing KpnI restriction site (Mir-181b-LUC-KpnI) were designed according to the sequence (from −1,316 to −1,183 bp relative to transcription start site) of human miR-181b promoter (Table S1). The amplified fragments were digested with KpnI restriction enzyme and inserted into pGL3-basic vector (Promega, USA). The promoter constructs were sequenced and incubated overnight with three units of CpG methyltransferase in the presence of 1 mmol/L S-adenosylmethionine according to the manufacture’s recommendation (New England Biolabs, UK). Then 1 µg of pGL3-basic (containing methylated or unmethylated miR-181b fragments) and 100 ng of Renilla plasmid (Promega, USA) were co-transfected into HEK293T using Lipofectamine 2000 for 24 hours. Luciferase and Renilla signals were measured 48 hours after transfection using Dual-Luciferase reporter assay kit (Promega, USA). The luciferase activity was normalized against Renilla activity. The experiments were performed in triplicate for each construct.
RNA isolation and quantitative RT-PCR
The isolation and purification of total RNA and quantitative RT-PCR was performed as previously described (20). Comparative cycle threshold (Ct) method was used to calculate expression of the miR-181b and the expression of U6 small nuclear RNA was considered as reference. The primer sequences predesigned for quantitative RT-PCR are tabulated in Table S1.
Western blots
The proteins from tissues and cells were obtained using a Cell Lysis extraction kit (Beyotime Biotechnology, Shanghai, China). The concentrations of the proteins were determined by using the BCA protein assay (Beyotime Biotechnology, China). Equal amounts (10 µg) of protein were constantly separated by proper concentrations of SDS-PAGE and transferred onto polyvinylidene difluoride membranes (PVDF, Millipore, USA). After blocked with 5% nonfat milk, PVDF membranes were incubated with the appropriate primary antibodies against PIAS1 (1:800; ab32219, Abcam, Cambridge, USA), KLF4 (1:1,000; ab215036, Abcam, Cambridge, USA), KLF6 (1:500; sc-134374, Santa Cruz, USA), YAP (1:500; sc-101199, Santa Cruz, USA), Fra-1 (1:500; sc-48424, Santa Cruz, USA) and GAPDH (1:2,000; 2118, Cell Signaling Technology, USA) at 4 °C for 16 hours. Then the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:2,500; catalog 7072, Cell Signaling Technology, USA) at room temperature for 2 hours. The bands were finally detected with ECL Detection Reagents (Thermo Fisher, USA).
Co-immunoprecipitation
The cells were lysed using a Cell Lysis extraction kit (Beyotime Biotechnology, Shanghai, China). After purification, the supernatants were obtained and incubated with the KLF4 antibody (1:100; ab215036, Abcam, Cambridge, USA) at 4 °C overnight. The immunoprecipitation was recovered by combination to protein A/G beads (Santa Cruz, USA) for 3 hours at 4 °C. After washing in lysing buffer, the immunoprecipitants were detected by immunoblot analysis.
Immunofluorescence staining
Macrophages transfected with miR-181b mimics and scramble mimics for 48 hours. The prepared cells were fixed with 3.6% paraformaldehyde for 10 minutes, permeabilized with 0.2% Triton X diluted in PBS (PBST), and subsequently blocked by treatment of cells with 3% bovine serum albumin (Beyotime Biotechnology, Shanghai, China). Then the cells were incubated with 5% normal goat serum diluted in PBST for 20 minutes at room temperature. CD206 antibodies (diluted 1:50, BeckmanCoulter, USA) were added and incubated at 4 °C for 16 hours and then the cells were labeled with goat-anti-mouse secondary antibodies coupled to cyanine 3 (Beyotime Biotechnology, Shanghai, China) at room temperature for 1 hour. Following appropriate washing steps with PBS and distilled water, nuclei were counterstained with DAPI (Beyotime Biotechnology, Shanghai, China).
Statistic analysis
Statistical analyses were performed with SPSS software (version 17.0, IBM, USA). The data were presented as mean ± standard deviation (SD). Differences between two group was analyzed with Student’s t-test. Differences between multiple groups were initially compared using one-way ANOVA test and then, if appropriate, between-group post-hoc tests were analyzed with Bonferroni comparison. Differences were regarded as statistically significance for P<0.05.
Results
MiR-181b is hypermethylated in peripheral monocytes from CAD patients
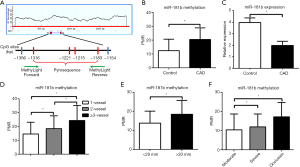
The characteristics of enrolled population is shown in Table 1. The locations of miR-181b promoter and CpG sites are shown in Figure 1A. The methylation status of miR-181b in circulating CD14+ monocytes isolated from CAD patients was remarkably higher than that from control group (control PMR 12.37±8.28 vs. CAD PMR 20.46±8.42, P<0.01, Figure 1B). By contrast, the miR-181b expression levels were greatly reduced in CAD patients relative to control group (Figure 1C). To further investigate the effect of miR-181b methylation on plaque characteristics, we stratified CAD patients by lesion length, the figure of culprit vessels and atherosclerotic burden. Interestingly, we found a gradual increase in the miR-181b methylation as increase in the number of culprit vessels, lesion length and the degree of stenosis (Figure 1D,E,F).
Full table
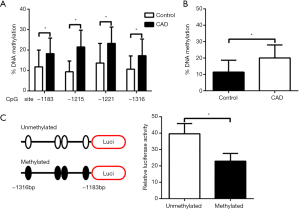
To further understand the precise methylation percentages within the promoter of miR-181b at indicated CpG sites, we next conducted pyrosequencing in CD14+ monocytes from 20 healthy controls and 20 CAD patients respectively. On basis of pyrosequencing, we accentuated that individual methylation percentage of 4 candidate CpG sites and the average methylation percentage of all analyzed CpG sites in CAD patients were consistently higher in CAD patients than those in control group (Figure 2A,B). In order to verify the function of methylation of CpG sites on miR-181b expression, we showed that as compared to normal vectors, the methylated vectors inserted with the indicated fragments of miR-181b promoter significantly repressed the transcription activities by 42.3% in HEK293T cells, reflecting that alteration in methylation status of miR-181b promoter is able to suppress miR-181b promoter activity and expression (Figure 2C).
Effect of miR-181b on macrophage polarization
We then sought to determine whether miR-181b affected macrophage polarization and activated inflammation. Result of quantitative RT-PCR showed that the expression levels of miR-181b were downregulated in macrophages induced by LPS and IFN-γ, while significant upregulation of miR-181b was observed in IL-4-treated macrophages (Figure 3A). To investigate the influence of miR-181b on macrophage polarization, we tested the expression of macrophage polarization markers by quantitative PCR. Overexpression of miR-181b significantly suppressed the expression of M1 phenotype markers such as IL-6 and TNF-α in macrophages stimulated by LPS and IFN-γ, but enhanced the expression of M2 markers such as Arg1 and CD206 in IL-4-induced macrophages (Figure 3B,C,D,E). Immunofluorescence staining of macrophages transfected with scramble or miR-181b mimics exhibited that the density and distribution of M2 phenotype marker CD206 was significantly increased in miR-181b-upregulated group as compared with scramble mimics group, implying that miR-181b promotes macrophages to polarize into M2 subset (Figure 3F).
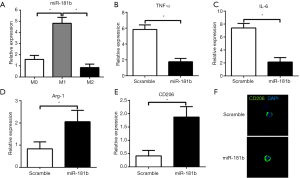
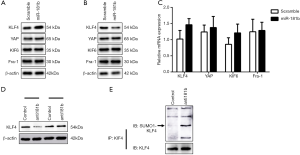
Effect of miR-181b on KLF4 expression and SUMOylation
To identify the potential molecular determinants involved in miR-181b-mediated macrophage polarization, we turned our attention to the effect of miR-181b on transcription factors priming macrophage polarization. Overexpression of miR-181b resulted in remarkable increase in the protein expression of KLF4 (Figure 4A). In contrast to KLF4 upregulation, we could not detect any induction of other known transcription factors in macrophages transfected with miR-181b mimics (Figure 4B). In fact, we failed to detect prominent discrepancy in the mRNA expression of KLF4 in macrophages transfected with scramble and miR-181b mimics (Figure 4C). This seeming contradiction inspired us to explore the post-transcriptional mechanism regulating miR-181b-induced KLF4 alteration. We pretreated macrophage with MG132 (10 µM), a potent proteasome inhibitor known to abrogate the levels of protein ubiquitylation and SUMOylation, finding that MG132 restored the reduced protein expression of KLF4 induced by miR-181b inhibition (Figure 4D). We then performed SUMOylation assay and observed an accumulation of SUMOylated KLF4 in macrophages transfected with anti181b, indicating that depletion of miR-181b augmented KLF4 SUMOylation and degradation (Figure 4E).
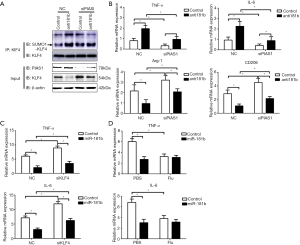
MiR-181b bound to PIAS1 and inhibited PIAS1 expression
To investigate the potential mechanisms involved in miR-181b-mediated KLF4 SUMOylation and M1 polarization, we searched for other putative target genes of miR-181b using online bioinformatic database, TargetScan Release 7.2 (www.targetscan.org). Among numerous conserved targets of miR-181b, PIAS1 was selected based on its role on protein degradation and SUMOylation. As shown in Figure 5A, there is a reserved binding sequence of miR-181b within the 3'UTR of PIAS1 mRNA. The binding fragment within the 3'UTR of PIAS1 was amplified and cloned into plasmid, followed with evaluation with Dual Luciferase Reporter Assay. The results showed that overexpression of miR-181b alleviated the luciferase activities of Luci-PIAS1-WT but did not alter the luciferase activities of Luci-PIAS1-Mut, suggesting that miR-181b was capable of directly binding to the 3'UTR of PIAS1 (Figure 5B). Furthermore, the results from quantitative PCR and immunoblots consistently showed that the expression of PIAS1 was dramatically decreased after up-regulation of miR-181b (Figure 5C,D). Accordingly, the expression levels of PIAS1 were markedly higher in circulating monocytes from CAD patients than those from healthy controls (Figure 5E). According to the prior report, we elucidated the effect of PIAS1 on activated STAT1 using Dual Luciferase assay, indicating that knockdown of PIAS1, in the presence of IFN-γ, resulted in increased Luciferase activity of STAT1 responsive elements (Figure 5F) (21).

MiR-181b silencing induced KLF4 SUMOylation via PIAS1 activation
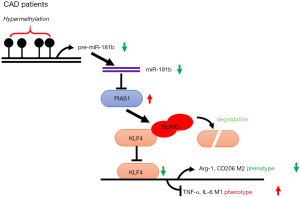
To further determine the relationship among miR-181b, PIAS1 and KLF4, the macrophages were co-transfected with miR-181b inhibitors and siPIAS1, followed by western blots, co-immunoprecipitation and quantitative PCR. Knockdown of miR-181b in macrophages enhanced PIAS1 and decreased the protein levels of KLF4, which were reversed by the downregulation of PIAS1 (Figure 6A). Consistent with the findings, knockdown of miR-181b in macrophages increased SUMO1-conjuageted KLF4 SUMOylation with this effect being counteracted by knockdown of PIAS1 (Figure 6A). The results of quantitative PCR showed that depletion of PIAS1 could counteract the suppressive effects of miR-181b inhibitors on Arg1 and CD206 expression, whereas supplements with siPIAS1 inhibited the excessive TNF-α and IL-6 expression induced by miR-181b inhibitors (Figure 6B). We next sought to determine whether miR-181b facilitated M1 polarized phenotype via regulating STAT1 or KLF4. As shown in Figure 6C, transfection with KLF4 siRNA substantially reversed the downregulation of TNF-α and IL-6 stimulated by overexpression of miR-181b. Nevertheless, specific STAT1 inhibitor Fludarabine did not affect the expression of TNF-α and IL-6 in the presence of miR-181b mimics (Figure 6D). These data suggested that miR-181b modulated macrophage polarization and abrogated KLF4 SUMOylation by interaction with PIAS1 (Figure 7).
Discussion
Recent studies have reported that miR-181b has critical function in numerous cancers, cardiac hypertrophy and kidney injury, but little is known about its roles in the progression of macrophage polarization (20,22-24). In this report, we underscore the predictive value of miR-181b hypermethylation in the incidence and development of CAD. Moreover, we provide a proof of principle for the genetic gain and loss of miR-181b regulating macrophage polarization via targeting PIAS1 and KLF4 SUMOylation.
The atherosclerotic microenvironmental milieu, which is composed of lipid metabolites, cytokines and non-coding RNAs secreted from parenchymal vascular cells and circulating immune cells, plays an important role in controlling macrophage polarization and in atherosclerotic development (4,6). In accordance to our results, Sun et al. (25) revealed that circulating miR-181b in the plasma was markedly reduced in CAD patients and in apolipoprotein E-deficient mice with a high-fat diet. However, the endogenous causation leading to the decline in miR-181b expression in CAD patients remains unclear. Toward this end, we performed MethyLight PCR and pyrosequencing, finding the specific methylated CpG sites at the promoter of miR-181b in CAD patients. A growing evidence of studies has uncovered alterations in DNA methylation at indicated promoters in specific cell types during the development of ASCVD (26). Among divergent types of circulating myeloid cells, monocytes are considered to play the most critical effect in the pathogenesis of atherosclerosis, because they are able to infiltrate into subendothelial, differentiate into pro-inflammatory M1 phenotype and anti-inflammatory M2 phenotype (5). Therefore, our findings propose that miR-181b hypermethylation in circulating monocytes may explain the decreased expression of miR-181b in CAD patients and become a promising marker for ASCVD.
It has been previously reported that increases in M1 phenotype and decreases in M2 phenotype are accompanied with the progression of atherosclerosis (27). Similar to the previous report, miR-181b was induced by LPS treatment and highly expressed in M1 subsets (28). The previous study had identified C/EBPβ as one of miR-181b target genes and confirmed that miR-181b inhibited M2 polarization via suppression of C/EBPβ (29). However, apart from C/EBPβ, many other lineage-specifying transcription factors, e.g., KLF4, KLF6, YAP and STAT1, have been shown to cooperatively govern macrophage polarization. While the mRNA expression of these transcription factors remained unchanged, the protein expression of KLF4 was prominently increased by overexpression of miR-181b. KLF4 acts as a critical transcription factor and centrally integrates multiple signal inputs in the pathogenesis of atherosclerosis (30,31). Li et al. (9) provided strong evidences that KLF4 retarded atherosclerotic progression via synergistically alleviating inflammation in endothelial cells and facilitating M1 to M2 phenotypic transition. In line with these results, we further accentuate that increased KLF4 by overexpression of miR-181b lead up to M2 phenotypic switch. By contrast, Wang et al. (32) observed that SUMOylation of KLF4 coordinated macrophages to polarize into M2 phenotype via enhancing the binding of KLF4 to Arg-1 upon IL-4 stimulation. This seemingly contradictory results could be explained by the fact that IL-4-induced KLF4 SUMOylation augmented KLF4 transcription activity but did not alter its expression and stability.
Another critical question is how miR-181b indirectly changes the protein expression of KLF4 via post-transcriptional modification. In the present study, delivery of miR-181b disturbs the M1 polarity switch through induction of KLF4 SUMOylation and degradation. In this regard, we show that knockdown of miR-181b exerts a facilitating action on M1 polarity via targeting PIAS1. Of note, PIAS1 belongs to the largest group of SUMO E3 ligase characterized by a SP-RING motif and is considered as the most critical one of E3 ligases (33). A number of researches has shown that PIAS1-linked SUMOylation of target transcription factor is engaged in diverse physiological progression and pathogenesis of numerous diseases (34,35). During the past two decades, several researchers have identified that the specific proteins were degraded by small ubiquitin-like modification (36). SUMOylation is referred to be a reversible procedure catalyzed by its own E1, E2 and E3 enzymes and is allowed to be restored by a diversity of SUMO-specific proteases (34,37). In line with the above observations, we demonstrated that knockdown of miR-181b augments KLF4 SUMOylation through promoting PIAS1 expression. Additionally, Kawai-Kowase et al. (38) delineated that PIAS1 induced KLF4 SUMOylation followed by proteasome degradation upon TGF-β stimulation in VSMCs. Given a direct modulation of miR-181b on PIAS1, it is likely that miR-181b has pleiotropic roles in the protein SUMOylation and in the pathogenesis of cardiovascular diseases.
Conclusions
Our data in clinical blood samples and molecular experiments highlight miR-181b as an epigenetic marker for atherosclerotic development and uncover its role in KLF4 SUMOylation and macrophage polarization via directly targeting PIAS1.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/cdt-20-407
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-407). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The clinical trial was approved by the First Affiliated Hospital of Harbin Medical University Ethics Committee (Ethical approval ID: HMU-2018-124). The informed written consent was provided in accordance with the Declaration of Helsinki (as revised in 2013) and obtained from the whole enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017;390:1151-210. [Crossref] [PubMed]
- Tabas I, Lichtman AH. Monocyte-Macrophages and T Cells in Atherosclerosis. Immunity 2017;47:621-34. [Crossref] [PubMed]
- Robbins CS, Hilgendorf I, Weber GF, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med 2013;19:1166-72. [Crossref] [PubMed]
- Groh L, Keating ST, Joosten LAB, et al. Monocyte and macrophage immunometabolism in atherosclerosis. Semin Immunopathol 2018;40:203-14. [Crossref] [PubMed]
- Murray PJ. Macrophage Polarization. Annu Rev Physiol 2017;79:541-66. [Crossref] [PubMed]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity 2010;32:593-604. [Crossref] [PubMed]
- Rougeot J, Torraca V, Zakrzewska A, et al. RNAseq Profiling of Leukocyte Populations in Zebrafish Larvae Reveals a cxcl11 Chemokine Gene as a Marker of Macrophage Polarization During Mycobacterial Infection. Front Immunol 2019;10:832. [Crossref] [PubMed]
- Lv LL, Feng Y, Wu M, et al. Exosomal miRNA-19b-3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury. Cell Death Differ 2020;27:210-26. [Crossref] [PubMed]
- Li Z, Martin M, Zhang J, et al. Kruppel-Like Factor 4 Regulation of Cholesterol-25-Hydroxylase and Liver X Receptor Mitigates Atherosclerosis Susceptibility. Circulation 2017;136:1315-30. [Crossref] [PubMed]
- Shankman LS, Gomez D, Cherepanova OA, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628-37. [Crossref] [PubMed]
- Stavri S, Simionescu M, Kardassis D, et al. Kruppel-like factor 4 synergizes with CREB to increase the activity of apolipoprotein E gene promoter in macrophages. Biochem Biophys Res Commun 2015;468:66-72. [Crossref] [PubMed]
- Essandoh K, Li Y, Huo J, et al. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016;46:122-31. [Crossref] [PubMed]
- Canfran-Duque A, Rotllan N, Zhang X, et al. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol Med 2017;9:1244-62. [Crossref] [PubMed]
- Ganta VC, Choi MH, Kutateladze A, et al. A MicroRNA93-Interferon Regulatory Factor-9-Immunoresponsive Gene-1-Itaconic Acid Pathway Modulates M2-Like Macrophage Polarization to Revascularize Ischemic Muscle. Circulation 2017;135:2403-25. [Crossref] [PubMed]
- Jiang D, Sun M, You L, et al. DNA methylation and hydroxymethylation are associated with the degree of coronary atherosclerosis in elderly patients with coronary heart disease. Life Sci 2019;224:241-8. [Crossref] [PubMed]
- Zhong W, Li B, Xu Y, et al. Hypermethylation of the Micro-RNA 145 Promoter Is the Key Regulator for NLRP3 Inflammasome-Induced Activation and Plaque Formation. JACC Basic Transl Sci 2018;3:604-24. [Crossref] [PubMed]
- Kolossvary M, Szilveszter B, Edes IF, et al. Comparison of Quantity of Coronary Atherosclerotic Plaques Detected by Computed Tomography Versus Angiography. Am J Cardiol 2016;117:1863-7. [Crossref] [PubMed]
- Ferguson JF, Hinkle CC, Mehta NN, et al. Translational studies of lipoprotein-associated phospholipase A(2) in inflammation and atherosclerosis. J Am Coll Cardiol 2012;59:764-72. [Crossref] [PubMed]
- Zhuang J, Peng W, Li H, et al. Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS One 2012;7:e47193. [Crossref] [PubMed]
- An Y, Chen XM, Yang Y, et al. LncRNA DLX6-AS1 promoted cancer cell proliferation and invasion by attenuating the endogenous function of miR-181b in pancreatic cancer. Cancer Cell Int 2018;18:143. [Crossref] [PubMed]
- Liu B, Liao J, Rao X, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A 1998;95:10626-31. [Crossref] [PubMed]
- Zhang X, Yu J, Zhao C, et al. MiR-181b-5p modulates chemosensitivity of glioma cells to temozolomide by targeting Bcl-2. Biomed Pharmacother 2019;109:2192-202. [Crossref] [PubMed]
- Indrieri A, Carrella S, Romano A, et al. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol Med 2019;11:e8734. [Crossref] [PubMed]
- Pang X, Feng G, Shang W, et al. Inhibition of lncRNA MEG3 Protects Renal Tubular From Hypoxia-Induced Kidney Injury in Acute Renal Allografts by Regulating miR-181b/TNF-α Signaling Pathway. J Cell Biochem 2019;120:12822-31. [Crossref] [PubMed]
- Sun X, He S, Wara AKM, et al. Systemic delivery of microRNA-181b inhibits nuclear factor-kappaB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ Res 2014;114:32-40. [Crossref] [PubMed]
- Duan L, Hu J, Xiong X, et al. The role of DNA methylation in coronary artery disease. Gene 2018;646:91-7. [Crossref] [PubMed]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 2018;233:6425-40. [Crossref] [PubMed]
- Zhang W, Shen X, Xie L, et al. MicroRNA-181b regulates endotoxin tolerance by targeting IL-6 in macrophage RAW264.7 cells. J Inflamm (Lond) 2015;12:18. [Crossref] [PubMed]
- Su R, Lin HS, Zhang XH, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene 2015;34:3226-39. [Crossref] [PubMed]
- Han YH, Kim HJ, Na H, et al. RORalpha Induces KLF4-Mediated M2 Polarization in the Liver Macrophages that Protect against Nonalcoholic Steatohepatitis. Cell Rep 2017;20:124-35. [Crossref] [PubMed]
- He M, Huang TS, Li S, et al. Atheroprotective Flow Upregulates ITPR3 (Inositol 1,4,5-Trisphosphate Receptor 3) in Vascular Endothelium via KLF4 (Kruppel-Like Factor 4)-Mediated Histone Modifications. Arterioscler Thromb Vasc Biol 2019;39:902-14. [Crossref] [PubMed]
- Wang K, Zhou W, Cai Q, et al. SUMOylation of KLF4 promotes IL-4 induced macrophage M2 polarization. Cell Cycle 2017;16:374-81. [Crossref] [PubMed]
- Rott R, Szargel R, Shani V, et al. SUMOylation and ubiquitination reciprocally regulate alpha-synuclein degradation and pathological aggregation. Proc Natl Acad Sci U S A 2017;114:13176-81. [Crossref] [PubMed]
- Jones MC, Fusi L, Higham JH, et al. Regulation of the SUMO pathway sensitizes differentiating human endometrial stromal cells to progesterone. Proc Natl Acad Sci U S A 2006;103:16272-7. [Crossref] [PubMed]
- Roukens MG, Alloul-Ramdhani M, Vertegaal AC, et al. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol Cell Biol 2008;28:2342-57. [Crossref] [PubMed]
- Chang SC, Ding JL. Ubiquitination and SUMOylation in the chronic inflammatory tumor microenvironment. Biochim Biophys Acta Rev Cancer 2018;1870:165-75.
- Gong L, Qi R, Li DW. Sumoylation Pathway as Potential Therapeutic Targets in Cancer. Curr Mol Med 2017;16:900-5. [Crossref] [PubMed]
- Kawai-Kowase K, Ohshima T, Matsui H, et al. PIAS1 mediates TGFbeta-induced SM alpha-actin gene expression through inhibition of KLF4 function-expression by protein sumoylation. Arterioscler Thromb Vasc Biol 2009;29:99-106. [Crossref] [PubMed]


