
Downregulation of long noncoding RNA SNHG6 rescued propofol-induced cytotoxicity in human induced pluripotent stem cell-derived cardiomyocytes
Introduction
Propofol (PPF) is one of the widely used anesthetic reagents for non-procedural sedation in intensive care units. Although rarely occurring, PPF infusion syndrome (PRIS) is potentially lethal, may cause severe side effects, such as hyperlipidemia, rhabdomyolysis, renal and cardiac failures (1-3). Unfortunately, due to its nature of rarity, the underlying genetic mechanisms of PRIS are largely unknown.
During past decades, limited investigations had used animal models or human patient-derived samples to study PRIS (4,5). Among them, human induced pluripotent stem cells (iPSCs) had been showing significantly clinical potentials, as they may differentiate into various human cells or organs, such as cardiomyocytes (6,7). Studies also demonstrated that human iPSC-derived cardiomyocytes (HiPSC-CMs) possessed similar cellular and molecular characteristics as those in in situ human cardiac cells (8,9). Specifically, a recent study showed that HiPSC-CMs responded to in vitro PPF exposure with similar cytotoxic characteristics as observed in human patients with PRIS (10).
Long non-coding RNAs (lncRNAs) are groups of long-length (>200 nucleotides), non-protein-coding RNA transcriptomes, that were recently discovered to play critical roles in almost all phases of human health and diseases (11-13). In cardiomyocytes, various studies had demonstrated that lncRNAs are actively involved in many aspects of cardiac development and pathophysiology (14-16). For example, a lncRNA named as cardiac apoptosis-related lncRNA (CARL) was reported to modulate cardiomyocyte apoptosis and mitochondrial fission through miR-539 and Prohibitin 2 (PHB2) (17). In addition, lncRNA of Urothelial Cancer Associated 1 (UCA1) was found to induce cardiomyocyte hypertrophy through competitively binding of miR-184/HOXA9 epigenetic axis (18). Moreover, lncRNA of H19 was discovered to suppress cardiomyocyte autophagy in diabetic cardiomyopathy (19).
Of many of the lncRNAs associated with human health or diseases, small nucleolar RNA host gene 6 (SNHG6) belongs to the super lncRNA family of SNHG and was found to be an active epigenetic transcriptome in various types of human cancers (20-22). However, there are only limited reports demonstrating functional mechanisms of SNHG6 in the cardiovascular system. Among them, it was shown that SNHG6 is likely to be aberrantly expressed in patients with atherosclerosis, and SNHG6 may modulate the progression and pathology of atherosclerotic cardiac tissues (23-25). However, the possible biological function of SNHG6 involved in the process of PPF-induced cardiac cytotoxicity has never been elucidated.
In this study, we established an in vitro PPF-induced cardiac cytotoxicity model by introducing PPF exposure to the in vitro culture of HiPSC-CMs. Then, we investigate the expression and function of SNHG6 during this process. The goal of this study is to explore the epigenetic underlying mechanisms of PRIS.
Methods
This study was approved by the Ethics Committee for Human & Clinical Research at the People’s Hospital of China Three Gorges University in Yichang, Hebei Province, China (Approval ID: CTGU20190455). All procedures were performed in accordance with the Declaration of Helsinki (2013 version). Informed consent was obtained from the patient for publication of this manuscript.
Induction of HiPSC-CMs
Induction of in vitro HiPSC-CMs was performed according to a previously published protocol with slight modification (26). A human embryonic stem cell (hESC) H7 cell line (Research Resource Identifier, RRID: CVCL_S799) was purchased from WiCell (WiCell Research Institute, USA) and maintained in 6-well tissue-culture plates in mTeSR medium (Stem Cell Technologies, Canada). For cardiac differentiation, H7 cells were moved into glass-bottomed dishes to form floating embryoid bodies in 48–72 h. Then, embryoid bodies were collected and re-plated in a 6-well tissue-culture plate in StemPro-34 medium (Thermo Fisher Scientific, USA) supplemented with 10 ng/mL BMP4 (MilliporeSigma, Shanghai, China), 2 mM glutamine (Thermo Fisher Scientific, USA), 4 mM monothioglycerol (MilliporeSigma, Shanghai, China), 50 µg/mL ascorbic acid (MilliporeSigma, Shanghai, China) and 3 ng/mL activin A (Thermo Fisher Scientific, USA) for 4 days. Then, 150 ng/mL DKK1 (Thermo Fisher Scientific) and 10 ng/mL VEGF were added into the culture for an additional 4 days. Finally, 5 ng/mL FGF-2 (MilliporeSigma, Shanghai, China) was added from day 9 and the medium was replenished every 3–4 days. From days 21 to 24, to purify HiPSC-CMs, beating cells were isolated and transferred to a new 6-well plate in RPMI medium (Thermo Fisher Scientific, USA) supplemented with non-essential amino acids (NEAA 1×, Thermo Fisher Scientific, USA), L-glutamine GlutaMAX (1×, Thermo Fisher Scientific, USA), antibiotic-antimycotic solution (1×, Thermo Fisher Scientific, USA) 4 mM lactic acid (MilliporeSigma, Shanghai, China) and 55 nM β-mercaptoethanol (Thermo Fisher, USA) for 7 days (27).
PPF treatment
In vitro treatment of PPF (MilliporeSigma, Shanghai, China) on HiPSC-CMs was conducted according to the method in a previously published study (10), with slight modification. Briefly, HiPSC-CMs were collected from 6-well plates and re-seeded in 96-well plates at approximately 3.5×103/well in culture medium supplemented with fetal bovine serum (FBS, 10%, MilliporeSigma, Shanghai, China) for 24 h. After that, culture medium was freshly replenished with 2% FBS, with the addition of PPF (MilliporeSigma, Shanghai, China) at 0, 1, 2, 5, 10, 20, 50 or 100 µg/mL for 48 h.
Cell viability assay
Cell viability assay was performed using a PrestoBlueTM HS Cell Viability Assay (Thermo Fisher Scientific, USA) according to the manufacturer’s instruction. Relative viability was assessed by measuring fluorescence (Excitation/Emission, 560/605) using a Fluoroskan microplate fluorometer (Thermo Fisher Scientific, USA).
Mitochondrial superoxide assay
The production of mitochondrial superoxide was assessed by immunofluorescence staining. To identify superoxide-induced mitochondrial oxidation, a MitoSOXTM Red Mitochondrial Superoxide Indicator dye (Thermo Fisher Scientific, USA) was applied and recognized by a TexasRed/mCherry filter cube on an Evos FL Auto fluorescent microscope system (Thermo Fisher Scientific, USA). In addition, cell nuclei were identified by using a Hoechst 33342 antibody (Thermo Fisher Scientific, Cat. # R37605, RRID: AB_2651135, USA) and recognized by a DAPI filter cube on the Evos FL Auto fluorescent microscope.
Mitochondrial membrane potential assay
A TMRE-Mitochondrial Membrane Potential Assay Kit (Thermo Fisher Scientific, USA) was used, according to the manufacturer’s instruction, to assess mitochondrial membrane potential. Th relative TRME signal was measured by fluorescence (Excitation/Emission, 530/580) using a Fluoroskan microplate fluorometer (Thermo Fisher Scientific, USA).
RNA extraction and quantitative real-time PCR (qRT-PCR)
RNA extraction and purification were conducted using a RNAqueousTM-Midi Total RNA Isolation Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instruction. The first-strand cDNA was then generated from RNA product using a SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, USA). Quantitative real-time PCR (qRT-PCR) was performed using an ABI Prism 7900 sequence detection system (Applied Biosystems, USA). For SNHG6, a pre-designed human TaqManTM SNHG6 Non-coding RNA Assay (Invitrogen, USA) was used. For hsa-miR-185-5p, a pre-designed hsa-miR-186-5p TaqMan® Advanced miRNA Assay (Applied Biosystems, USA) was used. Relative gene expression levels were characterized using the 2−∆∆Ct method.
SNHG6 downregulation assay
A lentiviral vector containing short-hairpin RNA (shRNA) specifically targeting human lncRNA SNHG6 (SNHG6_Sh), and another lentiviral vector containing a control non-specific shRNA (C_Sh) were designed and manufactured by GenePharma (GenePharma, Shanghai, China). Lentiviral packaging, transfection and supernatants collection/purification were also conducted by GenePharma (GenePharma, Shanghai, China). In the culture of H7 cells, cells were infected with lentiviral supernatants (SNHG6_Sh or C_Sh) for 48 h, followed by a selection process using G418 (0.5 mg/mL, MilliporeSigma, Shanghai, China) for 72 h. After 3–5 passages, cells were collected and examined by qRT-PCR to verify the downregulation of SNHG6. Then, these cells were differentiated toward HiPSC-CMs using the method described above.
Dual-luciferase reporter assay
The 3'-untranslated region (3'-UTR) of wild-type SNHG6, which includes a putative binding site of hsa-miR-186-5p was sub-cloned into pmiR-REPORT luciferase plasmid (Promega, USA) and named as SNHG6_Lu. In addition, the hsa-miR-186-5p binding site was mutated and the mutant SNHG6 3'-UTR was sub-cloned into pmiR-REPORT, named as SNHG6(MU)_Lu. In human HEK293T cells, SNHG6_Lu or SNHG6(MU)_Lu were co-transfected with hsa-miR-185-5p mimics (miR-186-5p, GenePharma, Shanghai, China) or a control non-specific human miRNA mimics (C_mmiC, GenePharma, Shanghai, China). Forty-eight hours later, a dual-luciferase reporter assay (Promega, USA) was performed. Relative Firefly/Renilla luciferase activities were then calculated and normalized to the luciferase activity in HEK293T cells co-transfected with SNHG_Lu and C_mmiC.
Hsa-miR-186-5p downregulation assay
A miRNA inhibitor specifically targeting hsa-miR-186-5p (miR186_In), and a control non-specific miRNA inhibitor (C_In) were both purchased from GenePharma (GenePharma, Shanghai, China). In the culture of purified HiPSC-CMs, cells were collected and re-seeded into 6-well plates in fresh culture medium with the addition of 10% FBS for 12 h. Transient transfection of miR186_In or C_In was then conducted for 12 h using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, USA). After that, qRT-PCR was performed to verify the downregulation of hsa-miR-186-5p.
Statistical analysis
All experiments were independently repeated at least three times. Data were shown as means ± standard error of the mean (SEM). Statistical analyses were performed using an unpaired two-tail student’s t-test on SPSS software (SPSS, IBM, RRID: SCR_002865, USA). The difference was termed as statistically significant if a P value was less than 5% (0.05).
Results
PPF induced cytotoxicity and upregulated SNHG6 in HiPSC-CMs
HiPSC-CMs were exposed to PPF for 48 h. After that, a viability assay indicated that, at concentrations between 2 and 100 µg/mL, PPF induced significant cell death (Figure 1A, *P<0.05). Also, an immunofluorescent assay showed that, 50 µg/mL PPF markedly increased mitochondrial superoxide production in HiPSC-CMs (Figure 1B). In addition, a TMRE assay demonstrated that mitochondrial membrane potentials in HiPSC-CMs were drastically decreased by 5–100 µg/mL PPF (Figure 1C, *P<0.05). Moreover, a qRT-PCR assay indicated that human lncRNA, SNHG6, was upregulated by 5–100 µg/mL PPF (Figure 1D, *P<0.05).
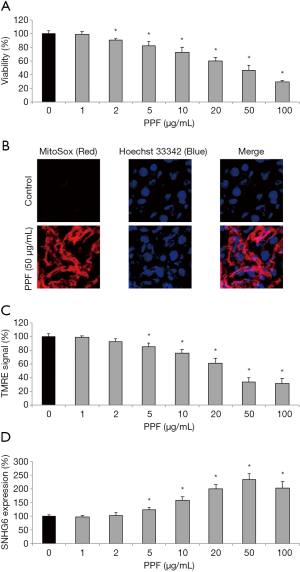
Thus, these data suggested that PPF induced significant cytotoxicity and upregulated SNHG6 in HiPSC-CMs.
SNHG6 downregulation rescued PPF-induced cytotoxicity in HiPSC-CMs
H7 cells were transduced with SNHG6-specific shRNA lentivirus (SNHG6_Sh) to knock down SNHG6 expression (Figure 2A, H7, *P<0.05). After cardiac differentiation, qRT-PCR assay confirmed that, SNHG6 was also downregulated in HiPSC-CMs differentiated from SNHG6_Sh-transduced H7 cells, as compared to those differentiated from H7 cells transduced with a control non-specific shRNA lentivirus (C_Sh) (Figure 2A, HiPSC-CMs, *P<0.05).
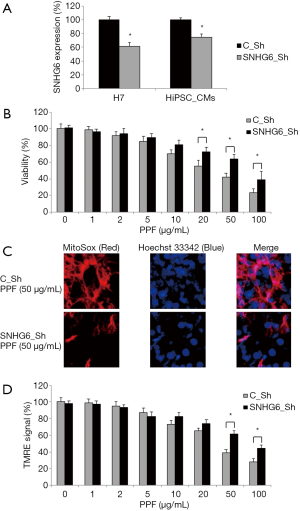
Then, transduced HiPSC-CMs were exposed to various concentrations of PPF (0–100 µg/mL) for 48 h, followed by a viability assay. The quantitative comparison demonstrated, while exposed to 20–100 µg/mL PPF, SNHG6_Sh-transduced HiPSC-CMs had much better surviving rates than C_Sh-transduced HiPSC-CMs (Figure 2B, *P<0.05).
Also, after PPF treatment, transduced HiPSC-CMs were examined by the fluorescent mitochondrial superoxide assay. It demonstrated that, while exposed to 50 µg/mL PPF, SNHG6_Sh-transduced HiPSC-CMs had much less mitochondrial superoxide production than C_Sh-transduced HiPSC-CMs (Figure 2C).
In addition, a TMRE assay indicated that, while exposed to 50–100 µg/mL PPF, SNHG6_Sh-transduced HiPSC-CMs had much less mitochondrial membrane potential drop than C_Sh-transduced HiPSC-CMs (Figure 2D, *P<0.05).
Thus, these data suggested that SNHG6 downregulation could rescue PPF-induced cytotoxicity in HiPSC-CMs.
Hsa-miR-186-5p is reversely correlated with SNHG16 expression in HiPSC-CMs
Using an online epigenetic targeting algorithm, StarBase 3.0 (28,29), it was noticed that human microRNA-186-5p (hsa-miR-186-5p) may serve as the downstream competing target of SNHG6 (Figure 3A). We thus constructed two luciferase vectors, one containing the wild-type human SNHG6 3'-UTR which hosts a putative hsa-miR-186-5p binding site (SNHG6_Lu), and the other containing the mutant SNHG6 3’-UTR with modified DNA sequences on the putative hsa-miR-186-5p binding site (SNHG6(MU)_Lu). Then, a dual-luciferase activity assay confirmed our hypothesis, indicating that SNHG6 could bind hsa-miR-186-5p (Figure 3B, *P<0.05, ∆P>0.05).
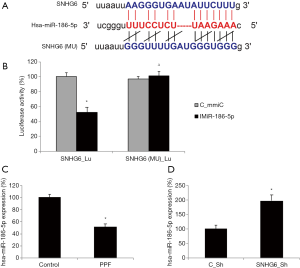
Also, we used qRT-PCR assay to investigate the expression pattern of hsa-miR-186-5p in HiPSC-CMs. It showed that, in HiPSC-CMs exposed to 50 µg/mL PPF, hsa-miR-186-5p expression was significantly lower than in control HiPSC-CMs (Figure 3C, *P<0.05). In addition, in lentiviral-transduced HiPSC-CMs, qRT-PCR indicated that SNHG6 downregulation inversely upregulated hsa-miR-186-5p (Figure 3D, *P<0.05).
Thus, these results suggested that hsa-miR-186-5p is the downstream competing target of SNHG6, thus reversely correlated with SNHG6 expression in HiPSC-CMs.
Hsa-miR-186-5p inhibition reversed the rescuing effect of SNHG6 downregulation on PPF-induced cytotoxicity in HiPSC-CMs
Lentiviral-transduced HiPSC-CMs were transiently transfected with hsa-miR-186-5p-targeted miRNA inhibitor (miR186_In) to knock down endogenous hsa-miR-186-5p expression (Figure 4A, *P<0.05). Along with those transfected with a control miRNA inhibitor (C_In), double-infected HiPSC-CMs were exposed to various concentrations of PPF (0–100 µg/mL) for 48 h. After that, the quantitative comparison demonstrated, while exposed to 5–50 µg/mL PPF, HiPSC-CMs double-infected with SNHG6_Sh/miR186_In had much higher death rates than HiPSC-CMs double-infected with SNHG6_Sh/C_In (Figure 4B, *P<0.05).
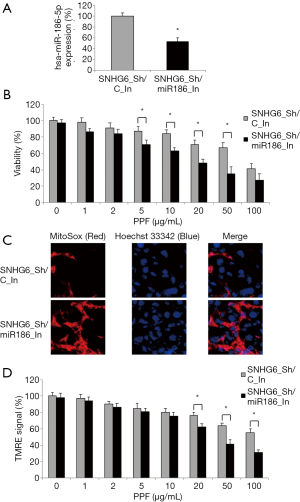
Also, the immunohistochemical assay showed that, while exposed to 50 µg/mL PPF, HiPSC-CMs double-infected with SNHG6_Sh/miR186_In had much more mitochondrial superoxide production than HiPSC-CMs double-infected with SNHG6_Sh/C_In (Figure 4C, *P<0.05).
Furthermore, a TMRE assay demonstrated that, while exposed to 20–100 µg/mL PPF, HiPSC-CMs double-infected with SNHG6_Sh/miR186_In had more mitochondrial membrane potential drops than HiPSC-CMs double-infected with SNHG6_Sh/C_In (Figure 4D, *P<0.05).
Therefore, these results indicated that hsa-miR-186-5p inhibition reversed the rescuing effect of SNHG6 downregulation on PPF-induced cytotoxicity in HiPSC-CMs.
Discussion
It was reported that, in order to prevent PRIS, the dosage of in vivo induction of PPF in human patients shall not be above 4 mg/kg/h for a 48-h time duration (1,2). In this study, we mimicked PPF-induced cardiac cytotoxicity in an in vitro model HiPSC-CMs. We discovered that cell viability was reduced and mitochondrial functions were damaged in HiPSC-CMs exposed to PPF approximately at a concentration of 5 µg/mL or higher. These findings are in line with the results in a previous study [10], and in accordance with the prediction that the non-toxic plasma concertation PPF was around 2.5 µg/mL (30).
Also in this study, we discovered SNHG6 was upregulated in PPF-exposed HiPSC-CMs. In a previous study, it was demonstrated that SNHG6 was aberrantly upregulated in murine fetal cardiac tissues with ventricular septal defect, and forced overexpression of SNHG6 inhibited proliferation and induced apoptosis in cardiomyocyte cell line P19 cells (23). In addition, it was noted SNHG6 was upregulated in cardiac tissues in patients with atherosclerosis, and knockdown of SNHG6 alleviated oxidative-stress induced cardiac apoptosis (24,25). Thus, our results are in line with those in previous studies, suggesting that aberrant expression (mostly likely upregulation/overexpression) of SNHG6 is very likely associated with pathological conditions in cardiovascular diseases.
The most important findings of our study were that, after we knocked down SNHG6 expression in HiPSC, the downregulation pattern of SNHG6 can be maintained until cardiac differentiation and we were able to show the rescuing effects of SNHG6 downregulation on PPF-induced cytotoxicity in HiPSC-CMs. Within the super-family of SNHG lncRNAs, newly emerged evidence suggested that, other members than SNHG6, such as SNHG1 and SNHG16 are also actively involved in the development and pathology of cardiomyocytes (31,32). Our preliminary data also showed gene expression changes of other SNHG super-family than SNHG6 (data not shown). However, after gene modification at stem cell stage, downregulating or upregulating patterns of other SNHG family members could not be maintained in HiPSC-CM stage. Thus, future investigations, possibily utilizing other in vivo or in vitro models with much feasible gene-editing capabilities, would further our understanding on the biological functions of SNHG family in regulating PPF-induced cardiac cytotoxicity.
Also, in our study, we explored the intrinsic signaling network of SNHG6 responsible for rescuing PPF-induced cytotoxicity in HiPSC-CMs. We demonstrated that hsa-miR-186-5p was an endogenous ceRNA candidate of SNHG6 (using dual-luciferase activity assay), and their expressions were inversely correlated in PPF-exposed HiPSC-CMs (using qRT-PCR assay). More importantly, we verified hsa-miR-186-5p was the functional downstream competing target of SNHG6, as suppressing hsa-miR-186-5p reversed the rescuing effects of SNHG6 downregulation on PPF-induced cytotoxicity in HiPSC-CMs. It is worth noting that, during the processes of SNHG6 regulating other human diseases (such as human cancers), SNHG6 was reported to be associated with various ceRNA candidates, probably depending on the sites or organs of diseases (22,33). Thus, more studies are required, in order to fully understand the associated signaling pathways of SNHG6 during the process of PRIS.
Acknowledgments
The authors thank Dr. Tianyu Qian for his critical review on the manuscript.
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/cdt-20-443
Conflicts in Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-443). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee for Human & Clinical Research at the People’s Hospital of China Three Gorges University in Yichang, Hebei Province, China (approval ID: CTGU20190455). All procedures were performed in accordance with the Declaration of Helsinki (2013 version). Informed consent was obtained from the patient for publication of this manuscript.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Krajčová A, Waldauf P, Andel M, et al. Propofol infusion syndrome: a structured review of experimental studies and 153 published case reports. Crit Care 2015;19:398. [Crossref] [PubMed]
- Fudickar A, Bein B. Propofol infusion syndrome: update of clinical manifestation and pathophysiology. Minerva Anestesiol 2009;75:339-44. [PubMed]
- Parke TJ, Stevens JE, Rice AS, et al. Metabolic acidosis and fatal myocardial failure after propofol infusion in children: five case reports. BMJ 1992;305:613-6. [Crossref] [PubMed]
- Bergamini C, Moruzzi N, Volta F, et al. Role of mitochondrial complex I and protective effect of CoQ10 supplementation in propofol induced cytotoxicity. J Bioenerg Biomembr 2016;48:413-23. [Crossref] [PubMed]
- Hsing CH, Chen YH, Chen CL, et al. Anesthetic propofol causes glycogen synthase kinase-3beta-regulated lysosomal/mitochondrial apoptosis in macrophages. Anesthesiology 2012;116:868-81. [Crossref] [PubMed]
- Gai H, Leung EL, Costantino PD, et al. Generation and characterization of functional cardiomyocytes using induced pluripotent stem cells derived from human fibroblasts. Cell Biol Int 2009;33:1184-93. [Crossref] [PubMed]
- Zhang J, Wilson GF, Soerens AG, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 2009;104:e30-41. [Crossref] [PubMed]
- Cohen JD, Babiarz JE, Abrams RM, et al. Use of human stem cell derived cardiomyocytes to examine sunitinib mediated cardiotoxicity and electrophysiological alterations. Toxicol Appl Pharmacol 2011;257:74-83. [Crossref] [PubMed]
- Ma J, Guo L, Fiene SJ, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol 2011;301:H2006-17. [Crossref] [PubMed]
- Kido K, Ito H, Yamamoto Y, et al. Cytotoxicity of propofol in human induced pluripotent stem cell-derived cardiomyocytes. J Anesth 2018;32:120-31. [Crossref] [PubMed]
- Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92:337-46. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Khalil AM, Rinn JL. RNA-protein interactions in human health and disease. Semin Cell Dev Biol 2011;22:359-65. [Crossref] [PubMed]
- Chang CP, Han P. Epigenetic and lncRNA regulation of cardiac pathophysiology. Biochim Biophys Acta 2016;1863:1767-71.
- Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res 2015;116:737-50. [Crossref] [PubMed]
- Shen S, Jiang H, Bei Y, et al. Long Non-Coding RNAs in Cardiac Remodeling. Cell Physiol Biochem 2017;41:1830-7. [Crossref] [PubMed]
- Wang K, Long B, Zhou LY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 2014;5:3596. [Crossref] [PubMed]
- Zhou G, Li C, Feng J, et al. lncRNA UCA1 Is a Novel Regulator in Cardiomyocyte Hypertrophy through Targeting the miR-184/HOXA9 Axis. Cardiorenal Med 2018;8:130-9. [Crossref] [PubMed]
- Zhuo C, Jiang R, Lin X, et al. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget 2017;8:1429-37. [Crossref] [PubMed]
- Cao C, Zhang T, Zhang D, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 2017;36:1112-22. [Crossref] [PubMed]
- Yan K, Tian J, Shi W, et al. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem 2017;42:999-1012. [Crossref] [PubMed]
- Xu M, Chen X, Lin K, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol 2019;12:3. [Crossref] [PubMed]
- Jiang Y, Zhuang J, Lin Y, et al. Long noncoding RNA SNHG6 contributes to ventricular septal defect formation via negative regulation of miR-101 and activation of Wnt/beta-catenin pathway. Pharmazie 2019;74:23-8. [PubMed]
- Shan H, Guo D, Zhang S, et al. SNHG6 modulates oxidized low-density lipoprotein-induced endothelial cells injury through miR-135a-5p/ROCK in atherosclerosis. Cell Biosci 2020;10:4. [Crossref] [PubMed]
- Wu Y, Zhang F, Li X, et al. Systematic analysis of lncRNA expression profiles and atherosclerosis-associated lncRNA-mRNA network revealing functional lncRNAs in carotid atherosclerotic rabbit models. Funct Integr Genomics 2020;20:103-15. [Crossref] [PubMed]
- Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 2008;453:524-8. [Crossref] [PubMed]
- Tan S, Tao Z, Loo S, et al. Non-viral vector based gene transfection with human induced pluripotent stem cells derived cardiomyocytes. Sci Rep 2019;9:14404. [Crossref] [PubMed]
- Yang JH, Li JH, Shao P, et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 2011;39:D202-9. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Marsh B, White M, Morton N, et al. Pharmacokinetic model driven infusion of propofol in children. Br J Anaesth 1991;67:41-8. [Crossref] [PubMed]
- Wang D, Lin B, Zhang W, et al. Up-regulation of SNHG16 induced by CTCF accelerates cardiac hypertrophy by targeting miR-182-5p/IGF1 axis. Cell Biol Int 2020;44:1426-35. [Crossref] [PubMed]
- Liang S, Ren K, Li B, et al. LncRNA SNHG1 alleviates hypoxia-reoxygenation-induced vascular endothelial cell injury as a competing endogenous RNA through the HIF-1alpha/VEGF signal pathway. Mol Cell Biochem 2020;465:1-11. [Crossref] [PubMed]
- Ruan J, Zheng L, Hu N, et al. Long noncoding RNA SNHG6 promotes osteosarcoma cell proliferation through regulating p21 and KLF2. Arch Biochem Biophys 2018;646:128-36. [Crossref] [PubMed]


